
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,083,224 – 17,083,325 |
| Length | 101 |
| Max. P | 0.905029 |

| Location | 17,083,224 – 17,083,317 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.35 |
| Shannon entropy | 0.61120 |
| G+C content | 0.47719 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -12.96 |
| Energy contribution | -11.90 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
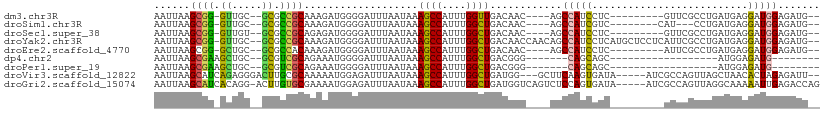
>dm3.chr3R 17083224 93 + 27905053 AAUUAAGCGG-GUUGC--GCGCCGCAAAGAUGGGGAUUUAAUAAAGCCAUUUGGUUGACAAC----AGCCAUCCUC---------GUUCGCCUGAUGAGGAUGGAGAUG-- ......((((-(....--.).))))..((((((.............)))))).........(----(.((((((((---------(((.....)))))))))))...))-- ( -27.22, z-score = -0.69, R) >droSim1.chr3R 23141661 91 - 27517382 AAUUAAGCGG-GUUGC--GCGCCGCAAAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAAC----AGCCAUCGUC--------CAU---CCUGAUGAGGAUGGAGAUG-- ......((((-(....--.).))))..((((((.............))))))(((((....)----))))(((.((--------(((---(((....))))))))))).-- ( -32.82, z-score = -2.71, R) >droSec1.super_38 354936 93 - 400794 AAUUAAGCGG-GUUGU--GCGCCGCAGAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAAC----AGCCAUCCUC---------GUUCGCCUGAUGAGGAUGGAGAUG-- ..........-(((((--...((.((....)).)).........((((....)))).)))))----..((((((((---------(((.....))))))))))).....-- ( -31.00, z-score = -1.72, R) >droYak2.chr3R 5874743 106 + 28832112 AAUUAAGCGG-GUUGC--GCGCCGCAAAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAACCAACAGCCAUCCUCAUGCUCCUCAUUCGCCUGAUGAGGAUGGAGAUG-- ......(..(-(((((--...((.((....)).)).........((((....))))).)))))..)..((((((((((((.........))...)))))))))).....-- ( -32.30, z-score = -0.81, R) >droEre2.scaffold_4770 13169563 93 - 17746568 AAUUAAGCGG-GCUGC--GCGCCACAAAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAAC----AGCCAUCCUC---------AUUCGCCUGAUGAGGAUGGAGAUG-- ......(((.-....)--))((((....(((((.............)))))))))......(----(.((((((((---------(((.....)))))))))))...))-- ( -28.62, z-score = -1.31, R) >dp4.chr2 5585621 76 + 30794189 AAUUAAGCGAAGCUGC--GCGUCGCAGAAAUGGGGAUUUAAUAAAGCCAUUUGGCUGACGGG-------CAGCAGC------------------AUGGAGAUG-------- ......((...(((((--.((((.((....))............((((....)))))))).)-------)))).))------------------.........-------- ( -27.10, z-score = -3.04, R) >droPer1.super_19 1302218 76 + 1869541 AAUUAAGCGAAGCUGC--GCGUCGCAGAAAUGGGGAUUUAAUAAAGCCAUUUGGCUGACGGG-------CAGCAGC------------------AUGGAGAUG-------- ......((...(((((--.((((.((....))............((((....)))))))).)-------)))).))------------------.........-------- ( -27.10, z-score = -3.04, R) >droVir3.scaffold_12822 2837738 101 - 4096053 AAUUAAGCAUCAGAGGGACUUGCGCAAAAAUGGAGAUUUAAUAAAGCCAUUUGGCUGAUGG---GCUUCAAGUGAUA-----AUCGCCAGUUAGCUAACACUAGAGAUU-- ......(((...........)))....((((((.............))))))(((((((.(---((.((....))..-----...))).))))))).............-- ( -17.12, z-score = 1.41, R) >droGri2.scaffold_15074 2600731 105 + 7742996 AAUUAAGCAUCACAGG-ACUUGUGCGAAAAUGGAGAUUUAAUAAAGCCAUUUGGCUGAUGGUCAGUCUCCAGUGAUA-----AUCGCCAGUUAGGCAAAAAUUGAGACCAG ........(((((.(.-(....).).....((((((((......((((....)))).......))))))))))))).-----...(((.....)))............... ( -25.32, z-score = -1.07, R) >consensus AAUUAAGCGG_GCUGC__GCGCCGCAAAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAAC____AGCCAUCCUC_________GUUCGCCUGAUGAGGAUGGAGAUG__ ......((((.((.....)).))))...................((((....))))............(((((..........................)))))....... (-12.96 = -11.90 + -1.05)


| Location | 17,083,224 – 17,083,317 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 67.35 |
| Shannon entropy | 0.61120 |
| G+C content | 0.47719 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -8.90 |
| Energy contribution | -8.41 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17083224 93 - 27905053 --CAUCUCCAUCCUCAUCAGGCGAAC---------GAGGAUGGCU----GUUGUCAACCAAAUGGCUUUAUUAAAUCCCCAUCUUUGCGGCGC--GCAAC-CCGCUUAAUU --......(((((((.((....))..---------)))))))(((----(((........))))))....................((((...--.....-))))...... ( -20.20, z-score = 0.07, R) >droSim1.chr3R 23141661 91 + 27517382 --CAUCUCCAUCCUCAUCAGG---AUG--------GACGAUGGCU----GUUGUCAGCCAAAUGGCUUUAUUAAAUCCCCAUCUUUGCGGCGC--GCAAC-CCGCUUAAUU --....((((((((....)))---)))--------)).(((((..----(((...((((....))))......)))..)))))...((((...--.....-))))...... ( -28.60, z-score = -2.59, R) >droSec1.super_38 354936 93 + 400794 --CAUCUCCAUCCUCAUCAGGCGAAC---------GAGGAUGGCU----GUUGUCAGCCAAAUGGCUUUAUUAAAUCCCCAUCUCUGCGGCGC--ACAAC-CCGCUUAAUU --.....((((((((.((....))..---------))))))))..----......((((....))))...................((((...--.....-))))...... ( -22.80, z-score = -0.78, R) >droYak2.chr3R 5874743 106 - 28832112 --CAUCUCCAUCCUCAUCAGGCGAAUGAGGAGCAUGAGGAUGGCUGUUGGUUGUCAGCCAAAUGGCUUUAUUAAAUCCCCAUCUUUGCGGCGC--GCAAC-CCGCUUAAUU --.....((((((((((...((.........))))))))))))..((.((((((.((((....))))..........((((....)).))...--)))))-).))...... ( -30.80, z-score = -0.28, R) >droEre2.scaffold_4770 13169563 93 + 17746568 --CAUCUCCAUCCUCAUCAGGCGAAU---------GAGGAUGGCU----GUUGUCAGCCAAAUGGCUUUAUUAAAUCCCCAUCUUUGUGGCGC--GCAGC-CCGCUUAAUU --.......(((((((((....).))---------))))))((((----((((((((((....))).............((....))))))).--)))))-)......... ( -26.60, z-score = -1.01, R) >dp4.chr2 5585621 76 - 30794189 --------CAUCUCCAU------------------GCUGCUG-------CCCGUCAGCCAAAUGGCUUUAUUAAAUCCCCAUUUCUGCGACGC--GCAGCUUCGCUUAAUU --------.........------------------((.((((-------(.((((.((.((((((.............))))))..)))))).--)))))...))...... ( -21.32, z-score = -3.11, R) >droPer1.super_19 1302218 76 - 1869541 --------CAUCUCCAU------------------GCUGCUG-------CCCGUCAGCCAAAUGGCUUUAUUAAAUCCCCAUUUCUGCGACGC--GCAGCUUCGCUUAAUU --------.........------------------((.((((-------(.((((.((.((((((.............))))))..)))))).--)))))...))...... ( -21.32, z-score = -3.11, R) >droVir3.scaffold_12822 2837738 101 + 4096053 --AAUCUCUAGUGUUAGCUAACUGGCGAU-----UAUCACUUGAAGC---CCAUCAGCCAAAUGGCUUUAUUAAAUCUCCAUUUUUGCGCAAGUCCCUCUGAUGCUUAAUU --.......((((((((.....(((.(((-----(......((((((---(............)))))))...)))).))).....((....))....))))))))..... ( -19.20, z-score = -0.66, R) >droGri2.scaffold_15074 2600731 105 - 7742996 CUGGUCUCAAUUUUUGCCUAACUGGCGAU-----UAUCACUGGAGACUGACCAUCAGCCAAAUGGCUUUAUUAAAUCUCCAUUUUCGCACAAGU-CCUGUGAUGCUUAAUU .............(((((.....))))).-----((((((((((((.........((((....))))........)))))).....((....))-...))))))....... ( -22.53, z-score = -0.99, R) >consensus __CAUCUCCAUCCUCAUCAGGCGAAC_________GAGGAUGGCU____GUUGUCAGCCAAAUGGCUUUAUUAAAUCCCCAUCUUUGCGGCGC__GCAAC_CCGCUUAAUU .......(((((..........................)))))............((((....))))...................((((.(.......).))))...... ( -8.90 = -8.41 + -0.49)



| Location | 17,083,233 – 17,083,325 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 65.11 |
| Shannon entropy | 0.65229 |
| G+C content | 0.51335 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -9.28 |
| Energy contribution | -9.56 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.645162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17083233 92 + 27905053 ----GGUUGCGCGCCGCAAAGAUGGGGAUUUAAUAAAGCCAUUUGGUUGACAAC----AGCCAUCCUC---------GUUCGCCUGAUGAGGAUGGAGAUGGCCAGCAG--- ----...(((...((.((....)).))..........((((((((((((....)----))))((((((---------(((.....)))))))))..)))))))..))).--- ( -31.20, z-score = -1.30, R) >droSim1.chr3R 23141670 90 - 27517382 ----GGUUGCGCGCCGCAAAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAAC----AGCCAUCGUC--------CAU---CCUGAUGAGGAUGGAGAUGGCCAGCAG--- ----.(((((...((.((....)).)).........((((....))))).))))----.((((((.((--------(((---(((....))))))))))))))......--- ( -35.60, z-score = -2.92, R) >droSec1.super_38 354945 92 - 400794 ----GGUUGUGCGCCGCAGAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAAC----AGCCAUCCUC---------GUUCGCCUGAUGAGGAUGGAGAUGGCCAGCAG--- ----.....(((.((.((....)).))..........((((((((((((....)----))))((((((---------(((.....)))))))))..)))))))..))).--- ( -34.40, z-score = -2.04, R) >droYak2.chr3R 5874752 105 + 28832112 ----GGUUGCGCGCCGCAAAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAACCAACAGCCAUCCUCAUGCUCCUCAUUCGCCUGAUGAGGAUGGAGAUGGUCAGCAG--- ----((((((...((.((....)).)).........((((....))))).)))))....((((((..(((.(((.(((......))).))).)))..))))))......--- ( -35.30, z-score = -1.21, R) >droEre2.scaffold_4770 13169572 92 - 17746568 ----GGCUGCGCGCCACAAAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAAC----AGCCAUCCUC---------AUUCGCCUGAUGAGGAUGGAGAUGGCCAGCAG--- ----.((((..(.(((......))).)..........((((((((((((....)----))))((((((---------(((.....)))))))))..)))))))))))..--- ( -36.10, z-score = -2.84, R) >dp4.chr2 5585630 75 + 30794189 ---AAGCUGCGCGUCGCAGAAAUGGGGAUUUAAUAAAGCCAUUUGGCUGACGGG-------------------------------CAGCAGCAUGGAGAUGAGCAGCAG--- ---..(((((.((((.((....))............((((....)))))))).)-------------------------------)))).((.((........))))..--- ( -26.80, z-score = -2.44, R) >droPer1.super_19 1302227 75 + 1869541 ---AAGCUGCGCGUCGCAGAAAUGGGGAUUUAAUAAAGCCAUUUGGCUGACGGG-------------------------------CAGCAGCAUGGAGAUGAGCAGCAG--- ---..(((((.((((.((....))............((((....)))))))).)-------------------------------)))).((.((........))))..--- ( -26.80, z-score = -2.44, R) >droVir3.scaffold_12822 2837747 100 - 4096053 UCAGAGGGACUUGC-GCAAAAAUGGAGAUUUAAUAAAGCCAUUUGGCUGAUGG---GCUUCAAGUGAUAA-----UCGCCAGUUAGCUAACACUAGAGAUUGCGGUCAG--- .......(((...(-((((((((((.............))))))(((((((.(---((.((....))...-----..))).)))))))...........))))))))..--- ( -23.12, z-score = 0.02, R) >droGri2.scaffold_15074 2600740 105 + 7742996 UCACAGG-ACUUGU-GCGAAAAUGGAGAUUUAAUAAAGCCAUUUGGCUGAUGGUCAGUCUCCAGUGAUAA-----UCGCCAGUUAGGCAAAAAUUGAGACCAGGGCUGUCAC ..((((.-.((((.-.(.....((((((((......((((....)))).......)))))))).......-----..(((.....))).........)..)))).))))... ( -28.62, z-score = -1.07, R) >consensus ____GGUUGCGCGCCGCAAAGAUGGGGAUUUAAUAAAGCCAUUUGGCUGACAAC____AGCCAUCCUC_________GUUCGCCUGAUGAGGAUGGAGAUGGCCAGCAG___ .....((((.((.((.((....)).)).........((((....)))).....................................................))))))..... ( -9.28 = -9.56 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:46 2011