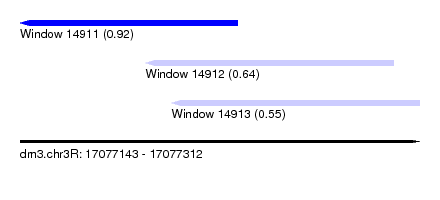
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,077,143 – 17,077,312 |
| Length | 169 |
| Max. P | 0.917418 |

| Location | 17,077,143 – 17,077,235 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.89 |
| Shannon entropy | 0.50920 |
| G+C content | 0.44765 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -15.09 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17077143 92 - 27905053 CCCAGAA-ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUUACUCGGCGUGUGGCUCAUUUGUCAGGGAUUG----GGAUUUGGACGA-ACUGGGACUU------------------ (((((..-.((((((..((((((..((.......))..))))))((((.(((...))).))))..))))))----(........)..-.)))))....------------------ ( -28.50, z-score = -2.19, R) >droSim1.chr3R 23135536 92 + 27517382 CCCAGAA-ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUUACUCGGCGUGUGGCUCAUUUGUCAGGGAUUG----GGAUUAGGAAGA-ACUGGGCCCU------------------ (((....-......((.((((((..((.......))..))))))..))..((((......)))))))...(----((.((((.....-.)))).))).------------------ ( -28.10, z-score = -1.75, R) >droSec1.super_38 348902 92 + 400794 CCCAGAA-ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUUACUCGGCGUGUGGCUCAUUUGUCAGGGAUUG----GGAUUAGGACGA-ACUGGGACUU------------------ (((((..-.((...((.((((((..((.......))..))))))..))..)).....((((((...(((..----..)))..)))))-))))))....------------------ ( -28.50, z-score = -2.36, R) >droYak2.chr3R 5868447 92 - 28832112 CCCAGAA-ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUUACUCGGCGUGUGGCUCAUUUGUCAGGGAUUG----GGAUUAGGACGC-ACUUGGAAUU------------------ .((((..-.........((((((..((.......))..)))))).((((.(..((((.((......)).))----))....).))))-..))))....------------------ ( -24.30, z-score = -1.12, R) >droEre2.scaffold_4770 13163551 92 + 17746568 CCCAGAA-ACAAUUCGAGAGUAAUGAUGAAGAAAGUUGUUACUCGGCGUGUGGCUCAUUUGUCAGGGAUUG----GGAUUAGGACGC-ACUGGGGCUU------------------ (((((..-.........((((((..((.......))..)))))).((((.(..((((.((......)).))----))....).))))-.)))))....------------------ ( -28.00, z-score = -1.91, R) >droAna3.scaffold_13340 3739233 95 - 23697760 CCCAGAA-ACAAUUUGAGAGUAAUGACGAAGAAAGUUGUUACUCGGCGUGUGGCUCAUUUGUCAGGGAUUG----GGAUUAGAGGAG-ACUGAGACCCAGG--------------- (((....-.........((((((..((.......))..))))))((((.(((...))).)))).))).(((----((.((..((...-.))..))))))).--------------- ( -29.70, z-score = -2.63, R) >dp4.chr2 5580102 77 - 30794189 CCCAAAA-ACCAUUUGAAAGUAAUGAUGAAGAAAGUUGUUACUUAGCAUGUGGCUCAUUUGUCAAGGAUUGUCGAGGA-------------------------------------- .((....-.((((.((.((((((..((.......))..))))))..)).)))).....(((.((.....)).))))).-------------------------------------- ( -14.60, z-score = -0.42, R) >droPer1.super_19 1296683 94 - 1869541 CCCAAAA-ACCAUUUGAAAGUAAUGAUGAAGAAAGUUGUUACUUAGCGUGUGGCUCAUUUGUCAAGGAUUGUCGAGGAUUGGCAGUGGGCCUGGG--------------------- ((((...-.........((((((..((.......))..)))))).......((((((((.(((((.............)))))))))))))))))--------------------- ( -26.42, z-score = -1.57, R) >droWil1.scaffold_181130 16053523 100 - 16660200 CCCAGAAUACAAUUUGAAAGUAAUGACGAAGAAAGUUGUUACUUAGCGUGUGGCUCAUUUGUCAGGAAUUG----GGAUUGGGAUUCUGAUGGGAGGGGGGCGG------------ (((....((((......((((((..((.......))..))))))....)))).(((.((..((((.((((.----.......))))))))..)).))))))...------------ ( -24.80, z-score = -1.24, R) >droMoj3.scaffold_6540 264435 115 + 34148556 CCCAGAA-ACAAUUUGAAGGUAAUGAGGAAGAAAGUUGUUACUUAGCUUGUGCCUCAUUUGUCAGCGAUUGCUCAGAGCUGAGAUGAGAAAAAGACAAAAAGAUGCUGAGAGACGA .......-.((((((((....(((((((....((((((.....))))))...)))))))..)))..)))))(((((..((....................))...)))))...... ( -23.85, z-score = -0.31, R) >droGri2.scaffold_15074 2594581 95 - 7742996 UCCAGAA-ACAAUUUGAACGUAAUGACGAAGAAAGUUGUUACUUAGCUUGUGGCUCAUUUGUCAGCGAUUGCUCAAAAGCAGGACAAGCAGACCUU-------------------- ...((.(-(((((((...(((....)))....)))))))).))..(((((((((......))).....(((((....))))).)))))).......-------------------- ( -21.50, z-score = -0.46, R) >consensus CCCAGAA_ACAAUUUGAGAGUAAUGACGAAGAAAGUUGUUACUCGGCGUGUGGCUCAUUUGUCAGGGAUUG____GGAUUAGGACGA_ACUGGGACUU__________________ .........((((((..((((((..((.......))..))))))......((((......)))).))))))............................................. (-15.09 = -14.74 + -0.35)
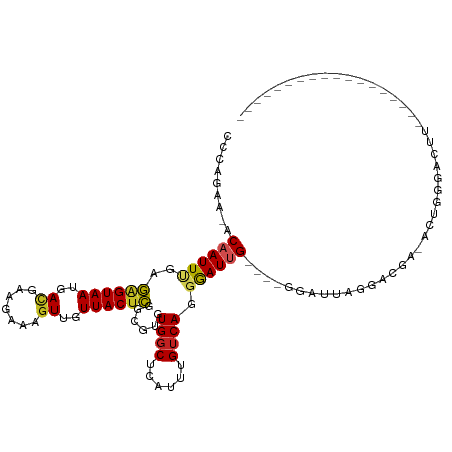
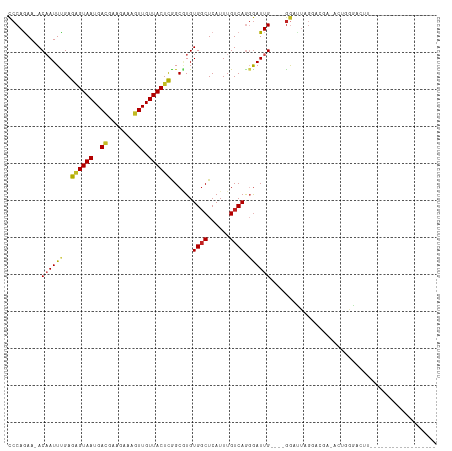
| Location | 17,077,196 – 17,077,301 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.34 |
| Shannon entropy | 0.45096 |
| G+C content | 0.44649 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -16.76 |
| Energy contribution | -18.19 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17077196 105 - 27905053 CAUUCAAUCCCAACCCUCUGCUCGCCAGGAUUGCGUGGGUGCUCACAGUUUUU-CAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUU ..........((((..(((((((....((((((..((((((((((.((((...-.)))).))).)))))))...-.))))))..))))........)))..)))).. ( -27.00, z-score = -1.02, R) >droSim1.chr3R 23135589 104 + 27517382 CAUACCAGCCC-AUCCUCCGCUCGCCAGGAUUGCGUGGGUGCUCACAGUUUUU-CAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUU .....((((.(-.((.((.((((....((((((..((((((((((.((((...-.)))).))).)))))))...-.))))))..))))...)).)).)...)))).. ( -26.50, z-score = -0.95, R) >droSec1.super_38 348955 88 + 400794 -----------------CCGCUCGCCAGGAUUGCGUGGGUGCUCAAAGUUUUU-CAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUU -----------------..((((....((((((..(((((((((((((((...-.)))))))).)))))))...-.))))))..))))((..((.......))..)) ( -29.50, z-score = -3.01, R) >droYak2.chr3R 5868500 104 - 28832112 CGGUUCACCCC-AUCCUCAGCUCACCAGGAUUGCGUGGGUGCUCACAGUUUUU-CAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUU .((......))-.((.(((((((....((((((..((((((((((.((((...-.)))).))).)))))))...-.))))))..))))..))).))........... ( -26.30, z-score = -0.63, R) >droEre2.scaffold_4770 13163604 103 + 17746568 CAGUUCAUCCC-AUCCUCCGCUCGCCUGGAUUGCGUGGGUGCUCACAG-UUUU-CAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGAUGAAGAAAGUUGUU ..........(-(((....((((...(((((((..((((((((((.((-(...-..))).))).)))))))...-.))))))).))))...))))............ ( -26.70, z-score = -0.86, R) >droAna3.scaffold_13340 3739289 102 - 23697760 CGGUCGGAGUC-CCCAUCAUCUCGACA--AUUGCGUGGGUGCUCACAGUUUUU-CAAUUUUGAUGCGCCCAGAA-ACAAUUUGAGAGUAAUGACGAAGAAAGUUGUU ..(((((....-.))....((((((..--((((..((((((((((.((((...-.)))).))).)))))))...-.)))))))))).....)))............. ( -25.90, z-score = -0.49, R) >droWil1.scaffold_181130 16053583 98 - 16660200 ------UGGUCUUCUUCCUUUUCAACA--AUUGCGUGGGUGCUCACAGUUUUU-CAAUUUUGAUGCGCCCAGAAUACAAUUUGAAAGUAAUGACGAAGAAAGUUGUU ------...(((((.((.(((((((..--((((..((((((((((.((((...-.)))).))).))))))).....)))))))))))....)).)))))........ ( -23.80, z-score = -1.26, R) >droVir3.scaffold_12822 2827569 87 + 4096053 -------------------GCAAGCCAAGCUUGUAUACGCUUACGUGUUUCUCACAAUUUGGAUGCGCCCAGAA-ACAAUUUGAACGUAAUGACGAAGAAAGUUGUU -------------------(((..(((((.(((((((((....))))).....))))))))).))).......(-(((((((...(((....)))....)))))))) ( -18.10, z-score = -0.25, R) >consensus C__U__A_CCC_ACCCUCCGCUCGCCAGGAUUGCGUGGGUGCUCACAGUUUUU_CAAUUUUGAUGCGCCCAGAA_ACAAUUCGAGAGUAAUGACGAAGAAAGUUGUU ...................((((....((((((..((((((((((.((((.....)))).))).))))))).....))))))..))))((..((.......))..)) (-16.76 = -18.19 + 1.42)

| Location | 17,077,207 – 17,077,312 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Shannon entropy | 0.44265 |
| G+C content | 0.46564 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -16.85 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17077207 105 - 27905053 GAUUCCCAUCCCAUUCAAUCCCAACCCUCUGC--UCGCCAGGAUUGCGUGGGUGCUCACAGUUUUUCAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGACGA .............................(((--((....((((((..((((((((((.((((....)))).))).)))))))...-.))))))..)))))....... ( -25.30, z-score = -1.05, R) >droSim1.chr3R 23135600 104 + 27517382 GGUUUCCUUCCCAUACCAGCCC-AUCCUCCGC--UCGCCAGGAUUGCGUGGGUGCUCACAGUUUUUCAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGACGA (((...........))).....-.......((--((....((((((..((((((((((.((((....)))).))).)))))))...-.))))))..))))........ ( -25.80, z-score = -0.56, R) >droYak2.chr3R 5868511 104 - 28832112 AAUCGUUUCAUCGGUUCACCCC-AUCCUCAGC--UCACCAGGAUUGCGUGGGUGCUCACAGUUUUUCAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGACGA ..(((((.....((......))-.......((--((....((((((..((((((((((.((((....)))).))).)))))))...-.))))))..))))...))))) ( -27.50, z-score = -1.27, R) >droEre2.scaffold_4770 13163615 102 + 17746568 -AUCGUUUCAGCAGUUCAUCCC-AUCCUCCGC--UCGCCUGGAUUGCGUGGGUGCUCACAG-UUUUCAAUUUUGAUGCGCCCAGAA-ACAAUUCGAGAGUAAUGAUGA -((((((((.((((((((....-.........--.....)))))))).((((((((((.((-(.....))).))).))))))))))-)).((((....).)))))).. ( -27.97, z-score = -1.08, R) >droAna3.scaffold_13340 3739300 98 - 23697760 --------AAUCAACCGGUCGGAGUCCCCAUC-AUCUCGACAAUUGCGUGGGUGCUCACAGUUUUUCAAUUUUGAUGCGCCCAGAA-ACAAUUUGAGAGUAAUGACGA --------.........(((((.....))...-.((((((..((((..((((((((((.((((....)))).))).)))))))...-.)))))))))).....))).. ( -25.90, z-score = -0.86, R) >droWil1.scaffold_181130 16053594 98 - 16660200 ----------AGAAGGCAUUUUGGUCUUCUUCCUUUUCAACAAUUGCGUGGGUGCUCACAGUUUUUCAAUUUUGAUGCGCCCAGAAUACAAUUUGAAAGUAAUGACGA ----------(((((((......)))))))...(((((((..((((..((((((((((.((((....)))).))).))))))).....)))))))))))......... ( -25.50, z-score = -1.62, R) >consensus _AU____UCACCAAUCCAUCCC_AUCCUCAGC__UCGCCAGGAUUGCGUGGGUGCUCACAGUUUUUCAAUUUUGAUGCGCCCAGAA_ACAAUUCGAGAGUAAUGACGA ..........................................(((((.((((((((((.((((....)))).))).)))))))(((.....)))....)))))..... (-16.85 = -16.80 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:43 2011