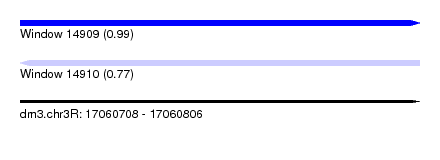
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,060,708 – 17,060,806 |
| Length | 98 |
| Max. P | 0.988790 |

| Location | 17,060,708 – 17,060,806 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 69.77 |
| Shannon entropy | 0.62111 |
| G+C content | 0.44182 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -14.14 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
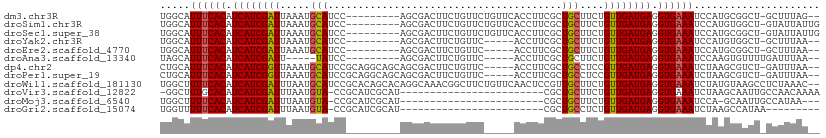
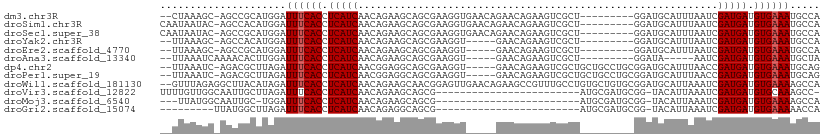
>dm3.chr3R 17060708 98 + 27905053 UGGCAUUUCACAUCAUCGAUUAAAUGCAUCC---------AGCGACUUCUGUUCUGUUCACCUUCGCUGCUUCUGUUGAUGAGGUGAAAUCCAUGCGGCU-GCUUUAG-- (((.(((((((.((((((((.....(((...---------.((((....((.......))...)))))))....)))))))).)))))))))).((....-)).....-- ( -25.80, z-score = -1.17, R) >droSim1.chr3R 23117873 100 - 27517382 UGGCAUUUCACAUCAUCGAUUAAAUGCAUCC---------AGCGACUUCUGUUCUGUUCACCUUCGCUGCUUCUGUUGAUGAGGUGAAAUCCAUGUGGCU-GUAUUAUUG (((.(((((((.((((((((.....(((...---------.((((....((.......))...)))))))....)))))))).)))))))))).......-......... ( -25.70, z-score = -1.37, R) >droSec1.super_38 331299 100 - 400794 UGGCAUUUCACAUCAUCGAUUAAAUGCAUCC---------AGCGACUUCUGUUCUGUUCACCUUCGCUGCUUCUGUUGAUGAGGUGAAAUCCAUGCGGCU-GUAUUAUUG (((.(((((((.((((((((.....(((...---------.((((....((.......))...)))))))....)))))))).)))))))))).......-......... ( -25.70, z-score = -1.25, R) >droYak2.chr3R 5844985 93 + 28832112 UGGCAUUUCACAUCAUCGAUUAAAUGCAUCC---------AGCGACUUCUGUUC-----ACCUUCGCUGCUUCUGUUGAUGAGGUGAAAUCCAUGUGGCU-GCUUUAA-- (((.(((((((.((((((((.....(((...---------.((((.........-----....)))))))....)))))))).)))))))))).......-.......-- ( -25.52, z-score = -1.69, R) >droEre2.scaffold_4770 13145460 93 - 17746568 UGGCAUUUCACAUCAUCGAUUAAAUGCAUCC---------AGCGACUUCUGUUC-----ACCUUCGCUGCUUCUGUUGAUGAGGUGAAAUCCAUGCGGCU-GCUUUAA-- (((.(((((((.((((((((.....(((...---------.((((.........-----....)))))))....)))))))).)))))))))).((....-)).....-- ( -25.62, z-score = -1.58, R) >droAna3.scaffold_13340 23528518 89 + 23697760 UAGCAUUUCACAUCAUCGAUU-----UAUCC---------AGCGACUUCUGUUC-----ACCUUCGCUGCUUCUGUUGAUGAGGUGAAAUCCAAGUGUUUUGAUUUAA-- ..(.(((((((.((((((((.-----.(..(---------(((((.........-----....))))))..)..)))))))).))))))).)................-- ( -24.82, z-score = -3.12, R) >dp4.chr2 5565018 102 + 30794189 CUGCAUUUCACAUCAUCGGUUAAAUGCAUCCGCAGGCAGCAGCGACUUCUGUUC-----ACCUUCGCUGCCUCCGUUGAUGAGGUGAAAUCUAAGCGUCU-GAUUUAA-- ..(((((((((.(((((((.....(((....)))((..(((((((.........-----....)))))))..)).))))))).)))))))....))....-.......-- ( -30.42, z-score = -1.80, R) >droPer1.super_19 1281637 102 + 1869541 CUGCAUUUCACAUCAUCGGUUAAAUGCAUCCGCAGGCAGCAGCGACUUCUGUUC-----ACCUUCGCUGCCUCCGUUGAUGAGGUGAAAUCUAAGCGUCU-GAUUUAA-- ..(((((((((.(((((((.....(((....)))((..(((((((.........-----....)))))))..)).))))))).)))))))....))....-.......-- ( -30.42, z-score = -1.80, R) >droWil1.scaffold_181130 16024538 108 + 16660200 UGGCUUUUCACAUCAUCGAUUUAAUGCAUCCGCACAGCACAGGCAAACGGCUUCUGUUCAACUCCGUUGCUUCUGUUGAUGAGGUGAAAUCUAUGUAAGCCUCUAAAC-- .((((((((((.(((((.......(((....))).((((.((((((.(((.............))))))))).))))))))).)))))........))))).......-- ( -29.42, z-score = -2.07, R) >droVir3.scaffold_12822 2806573 84 - 4096053 -GGCUUUGCACAUCAUCGAUUUAAUGUA-CCGCAUCGCAU------------------------CGCUGCUUCUGUUGAUGAGGUGAAAUCUAAGCAAUUGCCAACAAAA -(((.((((...((((((((.....(((-.((........------------------------)).)))....))))))))((......))..))))..)))....... ( -19.00, z-score = -0.33, R) >droMoj3.scaffold_6540 237330 81 - 34148556 UGGCUUUUCACAUCAUCGAUUUAAUGUA-CCGCAUCGCAU------------------------CGCUGCUUCUGUUGAUGAGGUGAAAUCCA-GCAAUUGCCAUAA--- (((..((((((.((((((((.....(((-.((........------------------------)).)))....)))))))).)))))).)))-((....)).....--- ( -21.20, z-score = -1.33, R) >droGri2.scaffold_15074 2582243 76 + 7742996 UGGUUUUUCACAUCAUCGAUUUAAUGUA-CCGCAUCGCAU------------------------CGCUGCCUCUGUUGAUGAGGUGAAAUCUAAGCCAUAA--------- (((((((((((.((((((((.....(((-.((........------------------------)).)))....)))))))).)))))....))))))...--------- ( -20.30, z-score = -2.04, R) >consensus UGGCAUUUCACAUCAUCGAUUAAAUGCAUCC_________AGCGACUUCUGUUC_____ACCUUCGCUGCUUCUGUUGAUGAGGUGAAAUCCAUGCGGCU_GCUUUAA__ .....((((((.((((((((.....(((............((((.....))))..............)))....)))))))).))))))..................... (-14.14 = -13.97 + -0.17)
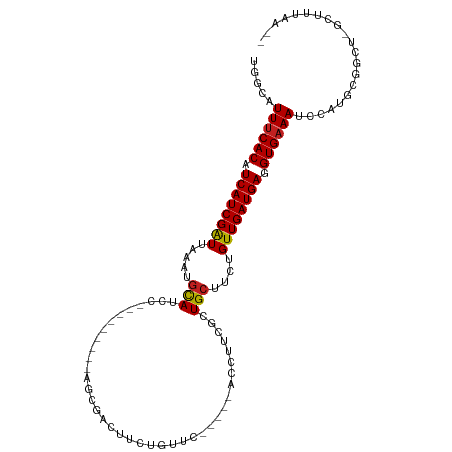
| Location | 17,060,708 – 17,060,806 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 69.77 |
| Shannon entropy | 0.62111 |
| G+C content | 0.44182 |
| Mean single sequence MFE | -26.81 |
| Consensus MFE | -8.58 |
| Energy contribution | -8.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
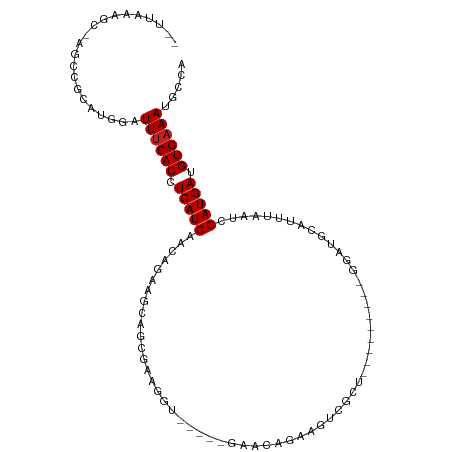
>dm3.chr3R 17060708 98 - 27905053 --CUAAAGC-AGCCGCAUGGAUUUCACCUCAUCAACAGAAGCAGCGAAGGUGAACAGAACAGAAGUCGCU---------GGAUGCAUUUAAUCGAUGAUGUGAAAUGCCA --.....((-....)).((((((((((.(((((........((((((..((.......)).....)))))---------)(((.......)))))))).))))))).))) ( -29.00, z-score = -2.13, R) >droSim1.chr3R 23117873 100 + 27517382 CAAUAAUAC-AGCCACAUGGAUUUCACCUCAUCAACAGAAGCAGCGAAGGUGAACAGAACAGAAGUCGCU---------GGAUGCAUUUAAUCGAUGAUGUGAAAUGCCA .........-.......((((((((((.(((((........((((((..((.......)).....)))))---------)(((.......)))))))).))))))).))) ( -27.80, z-score = -2.60, R) >droSec1.super_38 331299 100 + 400794 CAAUAAUAC-AGCCGCAUGGAUUUCACCUCAUCAACAGAAGCAGCGAAGGUGAACAGAACAGAAGUCGCU---------GGAUGCAUUUAAUCGAUGAUGUGAAAUGCCA .........-.......((((((((((.(((((........((((((..((.......)).....)))))---------)(((.......)))))))).))))))).))) ( -27.80, z-score = -2.22, R) >droYak2.chr3R 5844985 93 - 28832112 --UUAAAGC-AGCCACAUGGAUUUCACCUCAUCAACAGAAGCAGCGAAGGU-----GAACAGAAGUCGCU---------GGAUGCAUUUAAUCGAUGAUGUGAAAUGCCA --.......-.......((((((((((.(((((........((((((..(.-----...).....)))))---------)(((.......)))))))).))))))).))) ( -27.90, z-score = -2.49, R) >droEre2.scaffold_4770 13145460 93 + 17746568 --UUAAAGC-AGCCGCAUGGAUUUCACCUCAUCAACAGAAGCAGCGAAGGU-----GAACAGAAGUCGCU---------GGAUGCAUUUAAUCGAUGAUGUGAAAUGCCA --.....((-....)).((((((((((.(((((........((((((..(.-----...).....)))))---------)(((.......)))))))).))))))).))) ( -29.10, z-score = -2.49, R) >droAna3.scaffold_13340 23528518 89 - 23697760 --UUAAAUCAAAACACUUGGAUUUCACCUCAUCAACAGAAGCAGCGAAGGU-----GAACAGAAGUCGCU---------GGAUA-----AAUCGAUGAUGUGAAAUGCUA --................(.(((((((.(((((........((((((..(.-----...).....)))))---------)((..-----..))))))).))))))).).. ( -24.40, z-score = -2.91, R) >dp4.chr2 5565018 102 - 30794189 --UUAAAUC-AGACGCUUAGAUUUCACCUCAUCAACGGAGGCAGCGAAGGU-----GAACAGAAGUCGCUGCUGCCUGCGGAUGCAUUUAACCGAUGAUGUGAAAUGCAG --.......-....((....(((((((.(((((...((.((((((((..(.-----...).....)))))))).))(((....))).......))))).))))))))).. ( -33.60, z-score = -2.54, R) >droPer1.super_19 1281637 102 - 1869541 --UUAAAUC-AGACGCUUAGAUUUCACCUCAUCAACGGAGGCAGCGAAGGU-----GAACAGAAGUCGCUGCUGCCUGCGGAUGCAUUUAACCGAUGAUGUGAAAUGCAG --.......-....((....(((((((.(((((...((.((((((((..(.-----...).....)))))))).))(((....))).......))))).))))))))).. ( -33.60, z-score = -2.54, R) >droWil1.scaffold_181130 16024538 108 - 16660200 --GUUUAGAGGCUUACAUAGAUUUCACCUCAUCAACAGAAGCAACGGAGUUGAACAGAAGCCGUUUGCCUGUGCUGUGCGGAUGCAUUAAAUCGAUGAUGUGAAAAGCCA --.......(((((........(((((.(((((.((((..(((((((..((.....))..))).))))))))...((((....))))......))))).)))))))))). ( -31.60, z-score = -1.89, R) >droVir3.scaffold_12822 2806573 84 + 4096053 UUUUGUUGGCAAUUGCUUAGAUUUCACCUCAUCAACAGAAGCAGCG------------------------AUGCGAUGCGG-UACAUUAAAUCGAUGAUGUGCAAAGCC- ((((((((((....))...((.......))..))))))))(((...------------------------.)))...((.(-(((((((......))))))))...)).- ( -19.40, z-score = -0.14, R) >droMoj3.scaffold_6540 237330 81 + 34148556 ---UUAUGGCAAUUGC-UGGAUUUCACCUCAUCAACAGAAGCAGCG------------------------AUGCGAUGCGG-UACAUUAAAUCGAUGAUGUGAAAAGCCA ---....(((....))-)((.((((((.(((((.......(((...------------------------.)))((((...-..)))).....))))).))))))..)). ( -20.90, z-score = -0.98, R) >droGri2.scaffold_15074 2582243 76 - 7742996 ---------UUAUGGCUUAGAUUUCACCUCAUCAACAGAGGCAGCG------------------------AUGCGAUGCGG-UACAUUAAAUCGAUGAUGUGAAAAACCA ---------............((((((.(((((.......(((...------------------------.)))((((...-..)))).....))))).))))))..... ( -16.60, z-score = -0.68, R) >consensus __UUAAAGC_AGCCGCAUGGAUUUCACCUCAUCAACAGAAGCAGCGAAGGU_____GAACAGAAGUCGCU_________GGAUGCAUUUAAUCGAUGAUGUGAAAUGCCA .....................((((((.(((((............................................................))))).))))))..... ( -8.58 = -8.66 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:40 2011