
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,036,727 – 17,036,826 |
| Length | 99 |
| Max. P | 0.878516 |

| Location | 17,036,727 – 17,036,826 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.53581 |
| G+C content | 0.48088 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.48 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 17036727 99 - 27905053 ----------AAUGCAAAUGUGGGUUUUCACUUACUCCGCCACGUGUCCAGUUUGUGCUCUUCCAGCUGCUCCAUGGACGCAUUUGGCGAUUUGUAAUACAUUUGCAGG ----------..(((((((((((((........))))(((((.(((((((....(((((.....))).))....)))))))...))))).........))))))))).. ( -35.00, z-score = -3.43, R) >droSim1.chr3R 23091911 99 + 27517382 ----------AAUGCCAAUGUGUGUGUUCACUUACUUCGCCACGUGUCCAGUUUGUGCUCUUCCAGCUGCUCCAUGGACGCAUUUGGCGAUUUGUAAUACAUUUGCAGG ----------..(((.((((((((....)))((((.((((((.(((((((....(((((.....))).))....)))))))...))))))...)))).))))).))).. ( -34.10, z-score = -3.16, R) >droSec1.super_38 304618 99 + 400794 ----------AAUGCCAAUGUGUGUGUUCACUUACUUCGCCACGUGUCCAGUUUGUGCUCUUCCAGCUGCUCCAUGGACGCAUUUGGCGAUUUGUAAUACAUUUGCAGG ----------..(((.((((((((....)))((((.((((((.(((((((....(((((.....))).))....)))))))...))))))...)))).))))).))).. ( -34.10, z-score = -3.16, R) >droYak2.chr3R 5821570 98 - 28832112 -----------AUGCCAAUGUGUGUGUUCACUUACUUCGCCACGUGUCCAGUUUGUGCUCUUCCAGCUGCUCCAUGGACGCAUUUGGCGAUUUGUAAUACAUUUGCAGG -----------.(((.((((((((....)))((((.((((((.(((((((....(((((.....))).))....)))))))...))))))...)))).))))).))).. ( -34.10, z-score = -3.22, R) >droEre2.scaffold_4770 13122141 98 + 17746568 -----------AUGCCAAUGUGUGUGUUCACUUACUUCGCCACGUGUCCAGUUUGUGCUCUUCCAGCUGCUCCAUGGACGCAUUUGGCGAUUUGUAAUACAUUUGCAGG -----------.(((.((((((((....)))((((.((((((.(((((((....(((((.....))).))....)))))))...))))))...)))).))))).))).. ( -34.10, z-score = -3.22, R) >droAna3.scaffold_12911 2287073 95 - 5364042 -------------GAAAGCAUUAGUGUAU-CUUACUUCGCCACGUGUCCAGUUUGUGCUCUUCCAGCUGCUCCAUGGACGAAUUGGGCGAUUUGUAGUACAUCUGCAGG -------------....(((...((((((-..((..(((((...((((((....(((((.....))).))....)))))).....)))))..))..)))))).)))... ( -30.10, z-score = -2.02, R) >dp4.chr2 5303087 99 + 30794189 ----------CCACCUCAAGAUGUGUUCUACUCACUCCGCCACGUGUCCAGUUUGUGCUCUUCGAGCUGCUCCAUGGAGGCGUUGGGCGACUUGUAAUACAUUUGCAGG ----------...(((..((((((((..(((......((((((((.((((....((((((...)))).))....)))).))))..))))....)))))))))))..))) ( -32.00, z-score = -1.50, R) >droPer1.super_6 5322189 99 + 6141320 ----------CCACCUCAAGAUGUGUUCUACUUACUCCGCCACGUGUCCAGUUUGUGCUCUUCGAGCUGCUCCAUGGAGGCGUUGGGCGACUUGUAAUACAUUUGCAGG ----------...(((..((((((((..(((......((((((((.((((....((((((...)))).))....)))).))))..))))....)))))))))))..))) ( -32.00, z-score = -1.69, R) >droWil1.scaffold_181130 16003321 108 - 16660200 AUUUGAAUUUGAAUAAAGUUUUCA-AAGUACUCACUGCGCCAUGUAUCCAAUUUGUGCUCUUCUAGUUGCUCCAUAGAUGCAUUUGGGGUUUUAUAAUACAUUUGCAAG ...(((.((((((.......))))-))....))).((((..((((((((((...((((...((((.........)))).)))))))).........)))))).)))).. ( -17.10, z-score = 0.84, R) >droMoj3.scaffold_6540 21982221 95 - 34148556 --------------AUAUGGCUAAUGUAUACUGACUGCGCCACGUGUCCAGUUUGUGCUCCUCCAGCUGCUCCAUGGACGCGUUGGGUGACUUAUAAUACAUUUGCAAG --------------.....((.(((((((..(((...(((((((((((((....(((((.....))).))....)))))))))..))))..)))..))))))).))... ( -31.30, z-score = -2.25, R) >droGri2.scaffold_14906 4302598 96 + 14172833 ------------AGAUAU-GCAAAUAAAUACUUACUGCGCCACGUGUCCAAUUUGUGCUCCUCCAGCUGCUCCAUGGACGCAUUGGGUGACUUAUAAUACAUUUGCAAG ------------.....(-((((((............((((..(((((((....(((((.....))).))....)))))))....))))...........))))))).. ( -24.60, z-score = -1.54, R) >anoGam1.chr2R 57583765 92 + 62725911 -----------------CCCUUCACAGGUACUUACUUCGCCAAGUGUCCAGCUUGUGCUCCUCGAGCUCGUCCAUCGACGCGUUCGGUGAUUUGUAAUACAUUUUUAGA -----------------..((......(((.((((...(((((((.....))))).))((.((((((.((((....)))).)))))).))...)))))))......)). ( -24.10, z-score = -1.97, R) >triCas2.ChLG5 16430270 86 - 18847211 -----------------------CCUUCAACACGUGCCGCCAAGUGUCCAGUUUGUGCUCCUCAAGCUCGUCCAUCGCCACCGUUGGCGACUUGUAGUACAUUCGCAGG -----------------------(((.(.((((.((....)).))))...(..((((((...((((.(((.(((.((....)).)))))))))).))))))..)).))) ( -23.10, z-score = -1.44, R) >consensus ___________AUGCCAAUGUUUGUGUUUACUUACUUCGCCACGUGUCCAGUUUGUGCUCUUCCAGCUGCUCCAUGGACGCAUUUGGCGAUUUGUAAUACAUUUGCAGG .....................................((((..(((((........(((.....))).........)))))....)))).................... (-11.51 = -11.48 + -0.03)
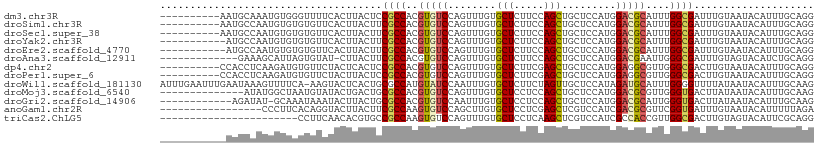

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:38 2011