
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,036,475 – 17,036,596 |
| Length | 121 |
| Max. P | 0.638241 |

| Location | 17,036,475 – 17,036,580 |
|---|---|
| Length | 105 |
| Sequences | 15 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.41 |
| Shannon entropy | 0.62904 |
| G+C content | 0.41330 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -13.24 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.46 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
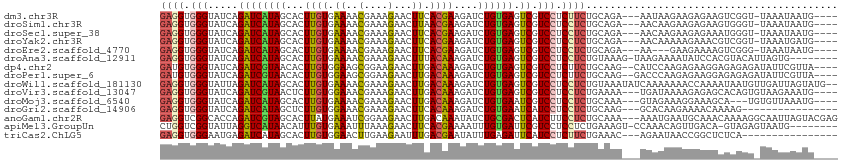
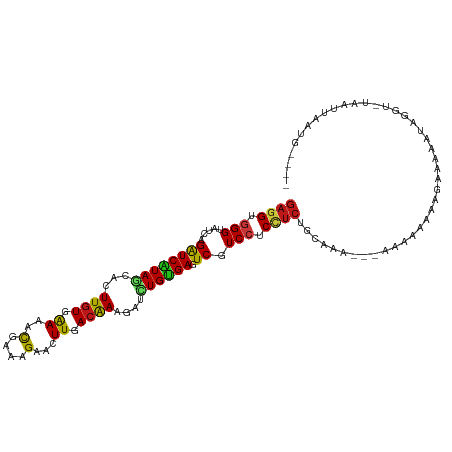
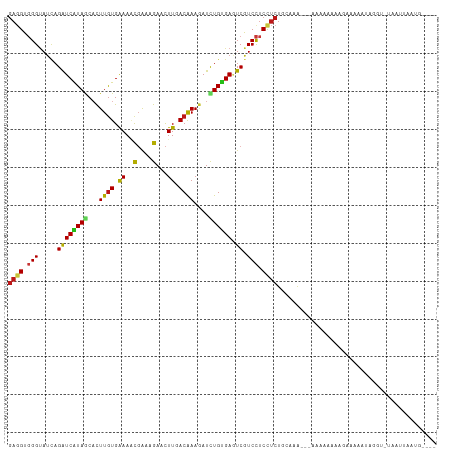
>dm3.chr3R 17036475 105 - 27905053 GAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUCACGAAGAUCUGUGAGUCGUCCUCUUCUGCAGA---AAUAAGAAGAGAAGUCGGU-UAAAUAAUG---- .((.(((.(....((((((((..((((((((..(....)...))))).)))..)))))).))...(((((((..(..---..).))))))).).))).)-)........---- ( -23.80, z-score = -0.49, R) >droSim1.chr3R 23091659 105 + 27517382 GAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUAACGAAGAUCUGUGAGUCGUCCUCCUCUGCAGA---AACAAGAAGAGAAGUGGGU-UAAAUAAUG---- (((((((.(..((((((....((....))....(....)...........))))))..).))).))))((((.(...---.....).))))........-.........---- ( -19.40, z-score = 0.70, R) >droSec1.super_38 304366 105 + 400794 GAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUCACGAAGAUCUGUGAGUCGUCCUCCUCUGCAGA---AACAAGAAGAGAAAUGGGU-UAAAUAAUG---- ((((.(((.....((((((((..((((((((..(....)...))))).)))..)))))).)).))).))))......---...................-.........---- ( -22.80, z-score = -0.23, R) >droYak2.chr3R 5821318 105 - 28832112 GAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUCACGAAGAUCUGUGAGUCGUCCUCCUCUGCAGA---AACAAAAAGAAACGUCGGU-UAAAUGAUG---- ((((.(((.....((((((((..((((((((..(....)...))))).)))..)))))).)).))).))))......---............(((((..-....)))))---- ( -25.40, z-score = -0.93, R) >droEre2.scaffold_4770 13121892 102 + 17746568 GAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUCACGAAGAUCUGUGAGUCGUCCUCCUCUGCAGA---AA---GAAGAAAAGUCGGG-UAAAUAAUG---- ((((.(((.....((((((((..((((((((..(....)...))))).)))..)))))).)).))).))))......---..---..............-.........---- ( -22.80, z-score = -0.36, R) >droAna3.scaffold_12911 2286822 104 - 5364042 GAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUUACAAAGAUCUGUGAGUCGUCCUCCUCUGUAAAG-UAAGAAAAUAUCCACGUACAUUAGUG-------- ...((((((((..((((((((...(((((((..(....)...)))))))....)))))).)).......(((......-..)))..))))))))...........-------- ( -22.80, z-score = -0.97, R) >dp4.chr2 5302833 107 + 30794189 GAUGUGGGUAUCAGAUCGUAACACUUGUGGAAGCGGAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCUUCUGCAAG--CAUCCAAGAGAAGGAGAGAGAUAUUCGUUA---- .....(((((((...((......((((..((((.(((.((.(((.(((......)))))))).))).))))..))))--..(((.......)))))..)))))))....---- ( -30.80, z-score = -1.07, R) >droPer1.super_6 5321935 107 + 6141320 GAUGUGGGUAUCAGAUCGUAACACUUGUGGAAGCGGAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCUUCUGCAAG--GACCCAAGAGAAGGAGAGAGAUAUUCGUUA---- .....(((((((...((......((((..((((.(((.((.(((.(((......)))))))).))).))))..))))--...((.......)).))..)))))))....---- ( -30.10, z-score = -0.81, R) >droWil1.scaffold_181130 16003063 111 - 16660200 GAGGUGGGUAUUAGAUCAUAGCACUUGUGAAAACGAAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCCUCUGUAAAUAUCAAAAAAACCAAAAUAAUGUUGAUUAGUAUG-- ((((.(((.....((((((((...((((.((..(....)...)).))))....)))))).)).))).)))).(((...(((((................)))))...))).-- ( -20.19, z-score = -0.68, R) >droVir3.scaffold_13047 14836373 106 + 19223366 GAGGUGGGUAUCAGAUCGUAACUCUUGUGGAAACGAAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCCUCUGAAAA---UGAUAAAAGAGAGCACAGUGUAAGAAAUG---- (((((((.(..((((((.(((.((((.((....)).))))..))).....))))))..).))).))))((((.....---.......))))..................---- ( -22.10, z-score = -0.14, R) >droMoj3.scaffold_6540 21981971 103 - 34148556 GAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUGACAAAGAUCUGUGAAUCGUCCUCCUCUGCAAA---GUAGAAAGGAAAGCA---UGUGUUAAAUG---- ((..((.....))..)).((((((.(((.....(((.(((.(((....))).))).....)))((((..(((((...---))))).))))..)))---.))))))....---- ( -20.90, z-score = -0.19, R) >droGri2.scaffold_14906 4302357 94 + 14172833 GAGGUGGGUAUCAGAUCAUAGCUCUUGUGGAAACGAAAGAACUUCACAAAGAUCUGUGAAUCAUCCUCCUCUGCAAG---GCACAAGAAAACAAAAG---------------- ((((.((((....((((((((.(((((((((..(....)...))))).)))).))))).))))))).))))......---.................---------------- ( -26.40, z-score = -2.48, R) >anoGam1.chr2R 57583508 110 + 62725911 GAGGUCGGCACCAGAUCGUAGCACUUAUGAAAUCGGAAGAACUUGACAAAUAUCUGCGACUCAUCUUCCUCUGCAAA---AAAUGAAUGCAAACAAAAGGCAAUUAGUACGAG ..(((....)))...(((((((.(((.((.....((((((..(((.((......)))))....))))))..((((..---.......))))..)).)))))......))))). ( -18.80, z-score = 0.48, R) >apiMel3.GroupUn 394601023 103 + 399230636 CUGGUCGGUAUUAGGUCAUAACAUUUGUGAAAUUUAAAGAACUUCACGAAAAUUUGUGAUUCGUCCUCCUCUGAAAGU-CCAAACAGUUGACA-GUAGAGUAAUG-------- ..((.(((......(((((((..((((((((.(((...))).))))))))...)))))))))).))..(((((...((-(.........))).-.))))).....-------- ( -20.30, z-score = -0.45, R) >triCas2.ChLG5 16428671 94 - 18847211 GAGGUGGGAAUGAGAUCAUAGCACUUGUGGAACUUGAAGAAUUUGACGAAUAUUUGAGAUUCAUCCUCUUCUGAAAC---AGAAUAACCGGCUCUCA---------------- (((((.((...(((.((((((...)))))).....((.((((((.(........).)))))).)))))(((((...)---))))...)).)).))).---------------- ( -18.40, z-score = 0.71, R) >consensus GAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCCUCUGCAAA___AAAAAAAAGAAAAAUAGGU_UAAUUAAUG____ ((((.(((.....((((((((...((((.((..(....)...)).))))....)))))).)).))).)))).......................................... (-13.24 = -12.85 + -0.38)
| Location | 17,036,501 – 17,036,596 |
|---|---|
| Length | 95 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Shannon entropy | 0.35155 |
| G+C content | 0.46142 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -19.23 |
| Energy contribution | -18.89 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
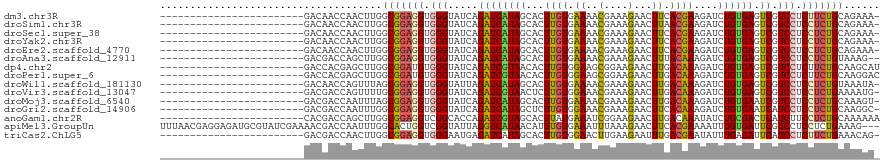
>dm3.chr3R 17036501 95 - 27905053 ------------------------GACAACCAACUUGGCGGAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUCACGAAGAUCUGUGAGUCGUCCUCUUCUGCAGAAA- ------------------------.............(((((((.(((.....((((((((..((((((((..(....)...))))).)))..)))))).)).))).))))))).....- ( -27.50, z-score = -1.32, R) >droSim1.chr3R 23091685 95 + 27517382 ------------------------GACAACCAACUUGGCGGAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUAACGAAGAUCUGUGAGUCGUCCUCCUCUGCAGAAA- ------------------------.............(((((((.(((.....((((((((..(((((.....(....)......)).)))..)))))).)).))).))))))).....- ( -25.10, z-score = -0.91, R) >droSec1.super_38 304392 95 + 400794 ------------------------GACAACCAACUUGGCGGAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUCACGAAGAUCUGUGAGUCGUCCUCCUCUGCAGAAA- ------------------------.............(((((((.(((.....((((((((..((((((((..(....)...))))).)))..)))))).)).))).))))))).....- ( -30.20, z-score = -2.09, R) >droYak2.chr3R 5821344 95 - 28832112 ------------------------GACAACCAACUUGGCGGAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUCACGAAGAUCUGUGAGUCGUCCUCCUCUGCAGAAA- ------------------------.............(((((((.(((.....((((((((..((((((((..(....)...))))).)))..)))))).)).))).))))))).....- ( -30.20, z-score = -2.09, R) >droEre2.scaffold_4770 13121915 95 + 17746568 ------------------------GACAACCAACUUGGCGGAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUCACGAAGAUCUGUGAGUCGUCCUCCUCUGCAGAAA- ------------------------.............(((((((.(((.....((((((((..((((((((..(....)...))))).)))..)))))).)).))).))))))).....- ( -30.20, z-score = -2.09, R) >droAna3.scaffold_12911 2286848 94 - 5364042 ------------------------GACGACCAGCUUGGCGGAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUUACAAAGAUCUGUGAGUCGUCCUCCUCUGUAAAG-- ------------------------.........(((.(((((((.(((.....((((((((...(((((((..(....)...)))))))....)))))).)).))).))))))).)))-- ( -27.10, z-score = -1.50, R) >dp4.chr2 5302860 96 + 30794189 ------------------------GACCACGAGCUUGGCGGAUGUGGGUAUCAGAUCGUAACACUUGUGGAAGCGGAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCUUCUGCAAGCAU ------------------------..(((((((....(((.((.((.....)).)))))....)))))))..((((((((...((((............))))...))))))))...... ( -29.50, z-score = -0.93, R) >droPer1.super_6 5321962 96 + 6141320 ------------------------GACCACGAGCUUGGCGGAUGUGGGUAUCAGAUCGUAACACUUGUGGAAGCGGAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCUUCUGCAAGGAC ------------------------..(((((((....(((.((.((.....)).)))))....)))))))..((((((((...((((............))))...))))))))...... ( -29.50, z-score = -0.88, R) >droWil1.scaffold_181130 16003095 95 - 16660200 ------------------------GACAACCAGUUUAGCGGAGGUGGGUAUUAGAUCAUAGCACUUGUGAAAACGAAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCCUCUGUAAAUA- ------------------------........(((((.((((((.(((.....((((((((...((((.((..(....)...)).))))....)))))).)).))).))))))))))).- ( -24.60, z-score = -1.32, R) >droVir3.scaffold_13047 14836400 95 + 19223366 ------------------------GACGACCAGUUUUGCGGAGGUGGGUAUCAGAUCGUAACUCUUGUGGAAACGAAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCCUCUGAAAAUG- ------------------------........(((((.((((((.(((...((((((.(((.((((.((....)).))))..))).....)))))).......))).))))))))))).- ( -25.80, z-score = -0.87, R) >droMoj3.scaffold_6540 21981995 95 - 34148556 ------------------------GACGACCAAUUUAGCGGAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUGACAAAGAUCUGUGAAUCGUCCUCCUCUGCAAAGU- ------------------------.............(((((((.(((.....((((((((...((((.((..(....)...)).))))....))))).))).))).))))))).....- ( -26.50, z-score = -1.82, R) >droGri2.scaffold_14906 4302372 95 + 14172833 ------------------------GACGACCAAUUUGGCGGAGGUGGGUAUCAGAUCAUAGCUCUUGUGGAAACGAAAGAACUUCACAAAGAUCUGUGAAUCAUCCUCCUCUGCAAGGC- ------------------------.............(((((((.((((....((((((((.(((((((((..(....)...))))).)))).))))).))))))).))))))).....- ( -33.80, z-score = -3.26, R) >anoGam1.chr2R 57583538 96 + 62725911 ------------------------CACGACCAGCUUGGCGGAGGUCGGCACCAGAUCGUAGCACUUAUGAAAUCGGAAGAACUUGACAAAUAUCUGCGACUCAUCUUCCUCUGCAAAAAA ------------------------.............(((((((..((....((.((((((...........((....)).............))))))))..))..)))))))...... ( -20.85, z-score = -0.06, R) >apiMel3.GroupUn 394601049 117 + 399230636 UUUAACGAGGAGAUGCGUAUCGAAAACGACCAAUUUGGCACUGGUCGGUAUUAGGUCAUAACAUUUGUGAAAUUUAAAGAACUUCACGAAAAUUUGUGAUUCGUCCUCCUCUGAAAG--- ......((((((..(((.........((((((.........))))))......((((((((..((((((((.(((...))).))))))))...))))))))))).))))))......--- ( -31.50, z-score = -2.37, R) >triCas2.ChLG5 16428686 95 - 18847211 ------------------------GACGACCAACUUGGCGGAGGUGGGAAUGAGAUCAUAGCACUUGUGGAACUUGAAGAAUUUGACGAAUAUUUGAGAUUCAUCCUCUUCUGAAACAG- ------------------------...........((.((((((.((((.((((.(((......((((.(((.........))).)))).....)))..))))))))))))))...)).- ( -18.00, z-score = 0.69, R) >consensus ________________________GACGACCAACUUGGCGGAGGUGGGUAUCAGAUCAUAGCACUUGUGAAAACGAAAGAACUUGACAAAGAUCUGUGAGUCGUCCUCCUCUGCAAAAA_ .....................................(((((((.(((.....((((((((...((((.((..(....)...)).))))....)))))).)).))).)))))))...... (-19.23 = -18.89 + -0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:37 2011