
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,998,112 – 16,998,228 |
| Length | 116 |
| Max. P | 0.752941 |

| Location | 16,998,112 – 16,998,228 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 73.40 |
| Shannon entropy | 0.49010 |
| G+C content | 0.40283 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16998112 116 + 27905053 CUUUACGACUAGAAGAAAAUAUUGUUUA-----GUUCCACUGCACGACUAAAUAAUAUUAAACACUAAAAGUCACAAGGCAUUUGUG-ACGCCAGUUUGUCUGGCACAGUCGGUCGCAGAUG .....(((((.......(((((((((((-----(((.........))))))))))))))...........((((((((...))))))-))(((((.....)))))..))))).......... ( -34.00, z-score = -3.58, R) >droEre2.scaffold_4770 13083536 113 - 17746568 CUUUACGACUAGAAGGAAGUAUUGUGUA-----GUUCCAAUGCACGACUAAAUAAUAUUAACCACUAAAAGUCACAAGACA---GAG-ACGCCAGUUUGUCUGGCAAAGUCGGCCGCAGAUG .....(((((....((.((((((((.((-----(((.........))))).))))))))..)).......(((....))).---...-..(((((.....)))))..))))).......... ( -26.20, z-score = -1.22, R) >droYak2.chr3R 5783938 118 + 28832112 CUUUAGUUCUAGAAGGAAAUAUUGUUUGAUUCGGUUCCAACGCACGACUAAAUAAAAUUAACCACUAAAAGUCACAAGGCA---GAA-ACGCCAGUUUGUCUGCCAAAGUCGGCCGCAGAUG .......(((....((((..(((....)))....))))...((.(((((..............((.....)).....((((---(((-((....)))..))))))..)))))))...))).. ( -22.40, z-score = 0.12, R) >droSec1.super_38 266858 113 - 400794 CUGUAAAUCUAGAAGAAAAUAUUGUUUA-----GUUCCACUGCAUGACUAAAUAAUAUUAACCACUAAAAGUCACAAGGCA---GAG-ACGCCAGUUUGUCUGGCACAGUCGGUCGCAGAUG ((((.............(((((((((((-----(((.........)))))))))))))).((((((....(((.(......---).)-))(((((.....)))))..))).))).))))... ( -29.10, z-score = -2.33, R) >droSim1.chr3R 23052644 113 - 27517382 CUUAAAAUCUAGAAGAAAAUAUUGUUUA-----GUUCCACUGCAUGACUAAAUAAUAUUAACCACUAAAAGUCACAAGGCA---GAG-ACGCCAGUUUGUCUGGCACAGUCGGCCGCAGAUG ......((((.......(((((((((((-----(((.........))))))))))))))..................(((.---..(-(((((((.....)))))...))).)))..)))). ( -28.80, z-score = -2.58, R) >droWil1.scaffold_181130 15959606 112 + 16660200 GACUAAAACUAGCAUCUAUGUUAGUUUG--------UCGCCGCAAAGUCUAGUCAGCUUAAGCAAUUGAUAUAUAUCUGCAUCUGUAUGUGUUAGCUGUAUCUGUACA-UCGGUAUCGGAU- (((..(((((((((....))))))))))--------))((((....((.(((.(((((...(((...(((....)))(((....))))))...)))))...))).)).-.)))).......- ( -26.50, z-score = -0.76, R) >consensus CUUUAAAACUAGAAGAAAAUAUUGUUUA_____GUUCCACUGCACGACUAAAUAAUAUUAACCACUAAAAGUCACAAGGCA___GAG_ACGCCAGUUUGUCUGGCACAGUCGGCCGCAGAUG .......................................((((.(((((.....................(((....)))..........(((((.....)))))..)))))...))))... (-13.21 = -13.88 + 0.67)

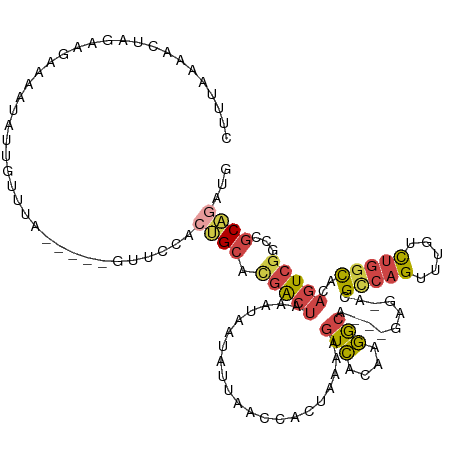
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:34 2011