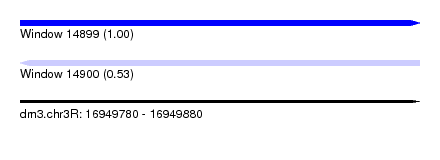
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,949,780 – 16,949,880 |
| Length | 100 |
| Max. P | 0.996000 |

| Location | 16,949,780 – 16,949,880 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.51 |
| Shannon entropy | 0.55548 |
| G+C content | 0.37684 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -12.82 |
| Energy contribution | -15.76 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.996000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
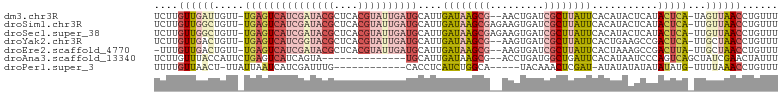
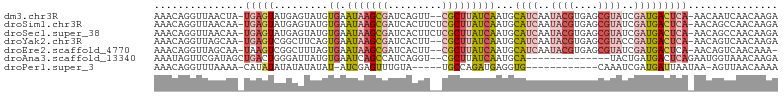
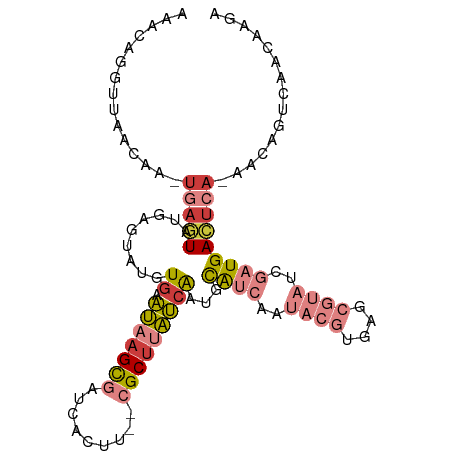
>dm3.chr3R 16949780 100 + 27905053 UCUUGUUGAUUGUU-UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCG--AACUGAUCGCUUAUUCACAUACUCAUACUCA-UAGUUAACCUGUUU ....(((((((((.-(((((((((((((((....))))))))))....((((((((--(.....))))))))).....))))).....)-))))))))...... ( -30.60, z-score = -4.14, R) >droSim1.chr3R 23009636 102 - 27517382 UCUUGUUGGCUGUU-UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCGAGAAGUGAUCGCUUAUUCACAUACUCAUACUCA-UUGUUAACCUGUUU ....((((((.((.-(((((((((((((((....))))))))))....(((((((((.......))))))))).....))))).))...-..))))))...... ( -31.20, z-score = -3.14, R) >droSec1.super_38 226219 102 - 400794 UCUUGUUGGCUGUU-UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCGAGAAGUGAUCGCUUAUUCACAUACUCAUACUCA-UUGUUAACCUGUUU ....((((((.((.-(((((((((((((((....))))))))))....(((((((((.......))))))))).....))))).))...-..))))))...... ( -31.20, z-score = -3.14, R) >droYak2.chr3R 5738969 100 + 28832112 UCUUGUUGACUGUU-UGAGUCAUCGGUACGCUCACGUAUUGAUGCAUUGAUAAGCG--AAGUGAUCGCUUAUUCACUGAAGCCGACUCA-UUGCUAACCUGUUU ....((((.(....-(((((((((((((((....))))))))).((.(((((((((--(.....))))))).))).)).....))))))-..).))))...... ( -29.50, z-score = -2.28, R) >droEre2.scaffold_4770 13042718 99 - 17746568 -UUUGUUGACUGUU-UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCG--AAGUGAUCGCUUAUUCACUAAAGCCGACUUA-UUGCUAACCUGUUU -...((((.(....-(((((((((((((((....))))))))).....((((((((--(.....)))))))))..........))))))-..).))))...... ( -26.50, z-score = -2.38, R) >droAna3.scaffold_13340 7875045 88 - 23697760 UCUUGUUUACCAUUCUGAGUCAUCAGUA--------------UGCAUUGAUAAGCG--ACCUGAUGGCUGAUUCACAUAAUCCCAGUCAGCUAUCGAACUAUUU .((((((...(((.((((....)))).)--------------))....))))))..--...((((((((((((...........))))))))))))........ ( -22.60, z-score = -2.47, R) >droPer1.super_3 5232005 84 - 7375914 UUUUGUUAACU-UUAUUAAUCAUCGAUUUG------------CACCUCAUCUGGCA-----UACAAACUCGAU-AUAUAUAUAUAUAUG-UUUUAAACCUGUUU ...........-.........(((((((((------------..((......))..-----..)))).)))))-((((((....)))))-)............. ( -7.50, z-score = -0.03, R) >consensus UCUUGUUGACUGUU_UGAGUCAUCGAUACGCUCACGUAUUGAUGCAUUGAUAAGCG__AAGUGAUCGCUUAUUCACAUAAUCAUACUCA_UUGUUAACCUGUUU ....((((((.....(((((((((((((((....))))))))))....((((((((.........))))))))...........)))))...))))))...... (-12.82 = -15.76 + 2.94)
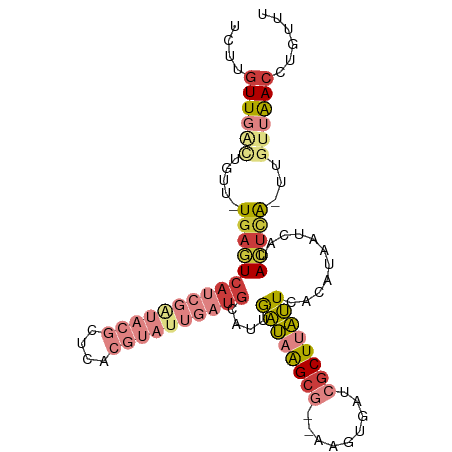
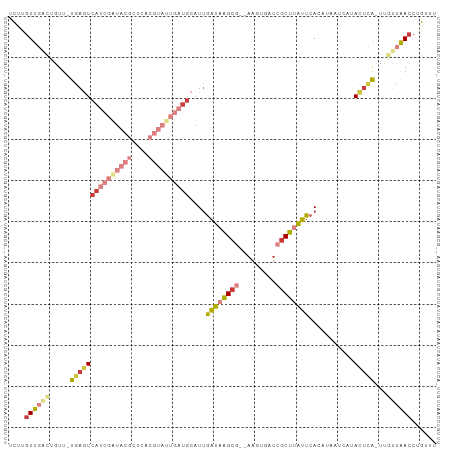
| Location | 16,949,780 – 16,949,880 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.51 |
| Shannon entropy | 0.55548 |
| G+C content | 0.37684 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -8.56 |
| Energy contribution | -10.11 |
| Covariance contribution | 1.55 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16949780 100 - 27905053 AAACAGGUUAACUA-UGAGUAUGAGUAUGUGAAUAAGCGAUCAGUU--CGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA-AACAAUCAACAAGA ..............-...((.(((((...(((.(((((((.....)--)))))))))...((((.(((((....))))).)))))))))-.))........... ( -26.90, z-score = -2.47, R) >droSim1.chr3R 23009636 102 + 27517382 AAACAGGUUAACAA-UGAGUAUGAGUAUGUGAAUAAGCGAUCACUUCUCGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA-AACAGCCAACAAGA .....((((.....-(((((....((((.(((.(((((((.......))))))))))))))(((.(((((....))))).))).)))))-...))))....... ( -27.20, z-score = -2.38, R) >droSec1.super_38 226219 102 + 400794 AAACAGGUUAACAA-UGAGUAUGAGUAUGUGAAUAAGCGAUCACUUCUCGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA-AACAGCCAACAAGA .....((((.....-(((((....((((.(((.(((((((.......))))))))))))))(((.(((((....))))).))).)))))-...))))....... ( -27.20, z-score = -2.38, R) >droYak2.chr3R 5738969 100 - 28832112 AAACAGGUUAGCAA-UGAGUCGGCUUCAGUGAAUAAGCGAUCACUU--CGCUUAUCAAUGCAUCAAUACGUGAGCGUACCGAUGACUCA-AACAGUCAACAAGA ......(((..(..-((((((.((.....(((.(((((((.....)--)))))))))..))(((..((((....))))..)))))))))-....)..))).... ( -27.10, z-score = -2.08, R) >droEre2.scaffold_4770 13042718 99 + 17746568 AAACAGGUUAGCAA-UAAGUCGGCUUUAGUGAAUAAGCGAUCACUU--CGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA-AACAGUCAACAAA- ..............-...((.((((....(((((((((((.....)--))))))).....((((.(((((....))))).))))..)))-...)))).))...- ( -24.80, z-score = -2.34, R) >droAna3.scaffold_13340 7875045 88 + 23697760 AAAUAGUUCGAUAGCUGACUGGGAUUAUGUGAAUCAGCCAUCAGGU--CGCUUAUCAAUGCA--------------UACUGAUGACUCAGAAUGGUAAACAAGA .....((((....(((((...............)))))((((((..--.((........)).--------------..)))))).....))))........... ( -16.76, z-score = 0.54, R) >droPer1.super_3 5232005 84 + 7375914 AAACAGGUUUAAAA-CAUAUAUAUAUAUAU-AUCGAGUUUGUA-----UGCCAGAUGAGGUG------------CAAAUCGAUGAUUAAUAA-AGUUAACAAAA ..............-..............(-(((((.(((((.-----.(((......))))------------))))))))))........-........... ( -11.70, z-score = -0.60, R) >consensus AAACAGGUUAACAA_UGAGUAUGAGUAUGUGAAUAAGCGAUCACUU__CGCUUAUCAAUGCAUCAAUACGUGAGCGUAUCGAUGACUCA_AACAGUCAACAAGA .....((((......(((((.........((.(((((((.........)))))))))...((((..((((....))))..)))))))))....))))....... ( -8.56 = -10.11 + 1.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:31 2011