
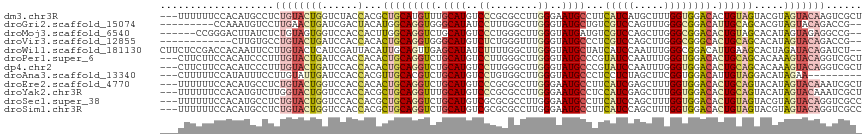
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,869,994 – 16,870,108 |
| Length | 114 |
| Max. P | 0.562748 |

| Location | 16,869,994 – 16,870,108 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Shannon entropy | 0.55569 |
| G+C content | 0.52448 |
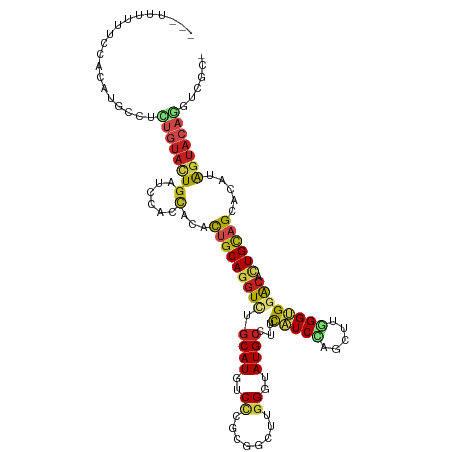
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -19.18 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16869994 114 - 27905053 ---UUUUUUCCACAUGCCUCUGUACUGGUCUACCACGCUGCAUGUUUGCAUGUCCCGCGCCUUGGGAAUGCCUUCAUCAUGCUUUGGUGGACACUGUAGUACGUAGUACAAGUCGCU ---............((..((((((((((((((((....(((((...((((.(((((.....)))))))))......)))))..))))))))((....))...)))))).))..)). ( -36.80, z-score = -2.05, R) >droGri2.scaffold_15074 2512681 106 + 7742996 ---------CCAAAUGUCCUUGAACUGAUCGACUACAUGGCAGGUGGGCAUAUCCUUUGGCUUGGGUAUGCUGUCGUCCAGUUUGGGCGGACAUUGCAGCACGUAGUACAGACCG-- ---------...((((((((..(((((..((((..(((.....)))(((((((((........)))))))))))))..)))))..)..)))))))....................-- ( -36.00, z-score = -1.06, R) >droMoj3.scaffold_6540 9139836 109 + 34148556 ------CCGGGACUUAUCUCUGUAGUGGUCCACCACUUGGCAGGUCUGCAUGUCCCUGGGCUUGGGUAUGAUGUCGUCCAGCUUGGGCGGACACUGUAGCACAUAGUAGAGGCCG-- ------.(((..(((....(((((((((....)))))..))))(((((((((((((..((((..((.(((....)))))))))..)..))))).))))).))......))).)))-- ( -38.50, z-score = 0.35, R) >droVir3.scaffold_12855 7649903 103 - 10161210 ------------CUUGUGCCUGUACUGAUCCACCACACUGCAGGUGGGCAUGUCUCUGGGUUUGGGUAUGCCCUCGUCCAGCUUGGGCGGGCACUGCAGCACAUAGUACAGACCG-- ------------.......((((((((......)...(((((((((((((((.(((.......))))))))))((((((.....)))))))).)))))).....)))))))....-- ( -39.30, z-score = -0.21, R) >droWil1.scaffold_181130 3238668 115 + 16660200 CUUCUCCGACCACAAUUCCUUGUACUCAUCGAUUACAUUGCAGUUGAGCAUAUCUUUUGGCUUGGGUAUGCUAUCAUCCAAUUUGGGCGGACAUUGAAGCACUAGAUACAGAUCU-- ((((((((.(((((((((..((((.((...)).))))..).)))))(((((((((........)))))))))...........))).))))....))))....((((....))))-- ( -26.30, z-score = -0.60, R) >droPer1.super_6 464466 114 + 6141320 ---CUUCUUCCACAUCCCUUUGUACUGAUCCACCACACUGCAGGUCUGCAUGUCCUUGGGCUUGGGUAUGCCCGUAUCCAAUUUGGGUGGACACUGCAGCACAAAGUACAGGUCGCU ---..............((((((.(((.(((((((((.((((....)))))))..(((((..((((....))))..)))))....)))))).....))).))))))........... ( -34.90, z-score = -0.44, R) >dp4.chr2 507919 114 + 30794189 ---CUUCUUCCACAUCCCUUUGUACUGAUCCACCACACUGCAGGUCUGCAUGUCCUUGGGCUUGGGUAUGCCCGUAUCCAAUUUGGGUGGACACUGCAGCACAAAGUACAGGUCGCU ---..............((((((.(((.(((((((((.((((....)))))))..(((((..((((....))))..)))))....)))))).....))).))))))........... ( -34.90, z-score = -0.44, R) >droAna3.scaffold_13340 7793519 105 + 23697760 ---CUUUUUCCAUAUUUCCUUGUAUUGAUCCACCACGUUGCACGUCUGCAUGUCCUGUGGCUUGGGUAUGCCCUCCUCUAGCUUCGGUGGACAUUGUAGGACAUAGAA--------- ---.........(((.((((.....((.((((((((((.(((....)))))))...(.((((.(((....)))......)))).)))))))))....)))).)))...--------- ( -28.10, z-score = -0.98, R) >droEre2.scaffold_4770 12961247 114 + 17746568 ---UUUUUUCCACAUGCCUCUGUACUGGUCCACCACACUGCAGGUCUGCAUGUCCCGCGCCUUGGGAAUGCCUUCAUCGAGCUUUGGUGGACACUGCAGUACAUAGUACAAAUCGCU ---............((...((((((((....))..((((((((((.((((.(((((.....)))))))))...((((((...))))))))).)))))))....))))))....)). ( -35.80, z-score = -1.60, R) >droYak2.chr3R 5676233 114 - 28832112 ---UUUUUUCCACAUGUCUUGGUACUGGUCCACCACGCUGCAGGUUUGCAUGUCCCGCGCCUUGGGAAUGCCUCCAUCGAGCUUUGGUGGACACUGCAGUACAUAGUACAAAUCGCU ---...........(((((..((((((((((((((.(((...((...((((.(((((.....)))))))))..))....)))..))))))))....))))))..)).)))....... ( -37.00, z-score = -1.41, R) >droSec1.super_38 144915 114 + 400794 ---UUUUUUCCACAUGCCUCUGUACUGGUCCACCACGCUGCAGGUCUGCAUGUCGCGCGCCUUGGGAAUGCCUUCAUCCAGCUUUGGUGGACACUGUAGUACGUAGUACAGGUCGCC ---............((.(((((((((((((((((.((((.((((.(((.....))).)))).(((....))).....))))..))))))))((....))...)))))))))..)). ( -38.40, z-score = -1.04, R) >droSim1.chr3R 22928517 114 + 27517382 ---UUUUUUCCACAUGCCUCUGUACUGGUCCACCACGCUGCAGGUCUGCAUGUCGCGCGCCUUGGGAAUGCCUUCAUCCAGCUUUGGUGGACACUGUAGUACGUAGUACAGGUCGCC ---............((.(((((((((((((((((.((((.((((.(((.....))).)))).(((....))).....))))..))))))))((....))...)))))))))..)). ( -38.40, z-score = -1.04, R) >consensus ___UUUUUUCCACAUGCCUCUGUACUGAUCCACCACACUGCAGGUCUGCAUGUCCCGCGGCUUGGGUAUGCCUUCAUCCAGCUUGGGUGGACACUGCAGCACAUAGUACAGGUCGC_ ...................((((((((......)...(((((((((.((((..((........))..))))...(((((.....)))))))).)))))).....)))))))...... (-19.18 = -19.68 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:28 2011