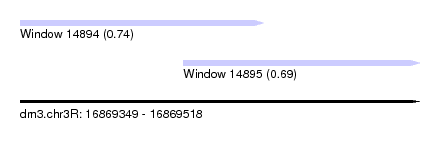
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,869,349 – 16,869,518 |
| Length | 169 |
| Max. P | 0.744470 |

| Location | 16,869,349 – 16,869,452 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.61 |
| Shannon entropy | 0.52473 |
| G+C content | 0.45902 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.54 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
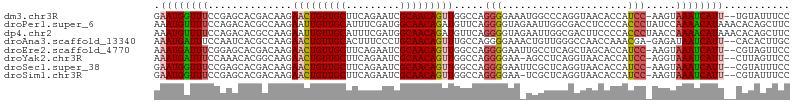
>dm3.chr3R 16869349 103 + 27905053 GAAUGGUUUCCGAGCACGACAAGAACUGUUGCUUCAGAAUCGCAACAGUUGGCCAGGGGAAAUGGCCCAGGUAACACCAUCC-AAGUAAAUCAUU--UGUAUUUCC (((((((((.(............(((((((((.........)))))))))(((((.......)))))..((.....))....-..).))))))))--)........ ( -27.20, z-score = -1.03, R) >droPer1.super_6 463850 106 - 6141320 AAAUGUUUUCCAGACACGCCAAGAAUUGUUGCAUUUCGAUGGCAACAGAUGUUCAGGGGUAGAAUUGGCGACCUCCCCACCCUAUCCAAAACAUAAACACAGCUUC ...(((((.........((((.(((.((...)).)))..)))).....(((((...((((((...(((.(....).)))..))))))..))))))))))....... ( -22.10, z-score = -0.19, R) >dp4.chr2 507306 106 - 30794189 AAAUGUUUUCCAGACACGCCAAGAAUUGUUGCAUUUCGAUGGCAACAGAUGUUCAGGGGUAGAAUUGGCGACUUCCCCACCCUAACCAAAACAUAAACACAGCUUC ..(((((((.............((((((((((.........))))))...)))).((((.((.........)).)))).........)))))))............ ( -19.50, z-score = 0.54, R) >droAna3.scaffold_13340 7792879 103 - 23697760 AAAUGAUUUCCAAUCACGCCAAGAACUGUUGCACUUUCCCUGCAACAGUUUGCCAGGGGAAACUGUUGGGCCAACCAAACGA-GAGAUAAUCAUU--CACACUUGC .(((((((..(..((.(.((..(((((((((((.......)))))))))))....)).)......((((.....))))..))-..)..)))))))--......... ( -27.60, z-score = -1.72, R) >droEre2.scaffold_4770 12960600 103 - 17746568 AAAUGAUUUCGGAGCACGACAAGAACUGUUGCUUCAGAAUCGCAACAGUUGGCCAGGGGAAUUGCCUCAGCUAGCACCAUCC-AAGUAAAUCAUU--CGUAGUUCC ..........(((((((((.......((((((.........))))))((((((..((((.....)))).)))))).......-...........)--))).))))) ( -28.10, z-score = -1.64, R) >droYak2.chr3R 5675581 102 + 28832112 AAAUGAUUUCCAAACACGGCAAGAACUGUUGCUUCAGAAUCGCAACAGUUGGCCAGGGGAA-AGCCUCAGGUAACACCAUCC-AGGUAAAUCAUU--CUUAGUUCC .((((((((((......(((...(((((((((.........))))))))).))).((((..-..)))).((.....))....-.)).))))))))--......... ( -31.40, z-score = -3.25, R) >droSec1.super_38 144270 103 - 400794 GAAUGGUUUCCGAGCACGACAAGAACUGUUGCUUCAGAAUCGCAACAGUUGGCCAGGGGAAUUCGCUCAGGUAACACCAUCC-AAGUAAAUCAUU--CGUAUUUCC (((((((((.(((((........(((((((((.........)))))))))..((...)).....)))).((.....))....-..).))))))))--)........ ( -28.30, z-score = -2.02, R) >droSim1.chr3R 22927873 102 - 27517382 GAAUGGUUUCCGAGCACGACAAGAACUGUUGCUUCAGAAUCGCAACAGUUGGCCAGGGGAA-UCGCUCAGGUAACACCAUCC-AAGUAAAUCAUU--CGUAUUUCC (((((((((.(((((........(((((((((.........)))))))))..((...))..-..)))).((.....))....-..).))))))))--)........ ( -28.30, z-score = -2.02, R) >consensus AAAUGAUUUCCAAGCACGACAAGAACUGUUGCUUCAGAAUCGCAACAGUUGGCCAGGGGAA_UGGCUCAGGUAACACCAUCC_AAGUAAAUCAUU__CGUAGUUCC .((((((((..............(((((((((.........))))))))).....((...................)).........))))))))........... (-13.36 = -13.54 + 0.17)
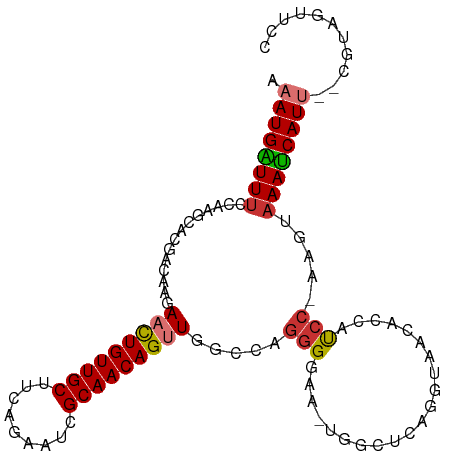
| Location | 16,869,418 – 16,869,518 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 62.07 |
| Shannon entropy | 0.77797 |
| G+C content | 0.43274 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -3.91 |
| Energy contribution | -3.82 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16869418 100 + 27905053 --GGUAACACCAUCCAAGUAAAUCAUU--UGUAUUUCCCAUAAUUUGUAACCCCUUUUUAUGGAA-ACUACAAGCGCAGGCGCGAUUAACAAGUGGUUGUAAACA----- --....(((((((....((.((((..(--((((.((.((((((...(......)...)))))).)-).)))))(((....))))))).))..)))).))).....----- ( -21.20, z-score = -1.24, R) >droPer1.super_6 463919 105 - 6141320 -GACCUCCCCACCCUAUCCAAAACAUAAACACAGCUUCUAUAAAAAAU-ACCCAUUUUUAUAGCA-ACCCCAAGCGCAGGCGCCUGCAACACGUGGCGGUAAAAUAUU-- -.....((((((.....................(((((((((((((..-.....)))))))))..-.....))))((((....)))).....)))).)).........-- ( -21.51, z-score = -3.12, R) >dp4.chr2 507375 105 - 30794189 -GACUUCCCCACCCUAACCAAAACAUAAACACAGCUUCUAUAAAAAAU-ACCCAUUUUUAUAGCA-ACCCCAAGCGCAGGCGCCUGCAACACGUGGCGGUAAAAUAAU-- -.....((((((.....................(((((((((((((..-.....)))))))))..-.....))))((((....)))).....)))).)).........-- ( -21.51, z-score = -3.03, R) >droAna3.scaffold_13340 7792948 102 - 23697760 --GCCAACCAAACGAGAGAUAAUCAUU--CACACUUGCCACAAUUUUGGACCCCUUUUUAUGGCA-GCCCUAGGCGCAGGCGCGCGCAACACGUGGCAAGAAAAUAA--- --...........(((.(.....).))--)...((((((((......((...((.......))..-.))....((((......)))).....)))))))).......--- ( -25.10, z-score = -0.67, R) >droEre2.scaffold_4770 12960669 101 - 17746568 --GCUAGCACCAUCCAAGUAAAUCAUU--CGUAGUUCCCAUAAUUUGUAGCCCCUUGUUAUGGAA-ACUGCAAGCGCAAGCGCGAUUAACAAGUGGUUGUAAACAA---- --....(((((((....((.((((...--.((((((.(((((((..(......)..))))))).)-)))))..((....))..)))).))..)))).)))......---- ( -28.80, z-score = -3.38, R) >droYak2.chr3R 5675649 102 + 28832112 --GGUAACACCAUCCAGGUAAAUCAUU--CUUAGUUCCCAUAAUUUGUAACCCCUUGUUAUGGAA-ACUCGAAGCGCAAGCGCGAUUAACAGGUGGUUGUCAACAAA--- --.((.((((((((...((.((((.((--(..((((.(((((((..(......)..))))))).)-))).)))((....))..)))).)).))))).)))..))...--- ( -27.30, z-score = -2.42, R) >droSec1.super_38 144339 100 - 400794 --GGUAACACCAUCCAAGUAAAUCAUU--CGUAUUUCCCAUAAUUUGUAACCCCUUUUUAUGGAA-ACUACAAGCGCAGGCGCGAUUAACAAGUGGUUGUAAACA----- --....(((((((....((.((((...--.(((.((.((((((...(......)...)))))).)-).)))..(((....))))))).))..)))).))).....----- ( -19.70, z-score = -0.95, R) >droSim1.chr3R 22927941 100 - 27517382 --GGUAACACCAUCCAAGUAAAUCAUU--CGUAUUUCCCAUAAUUUGGAACCCCUUUUUAUGGAA-ACUAAAAGCGCAGGCGCGAUUAACAAGUGGUUGUAAACA----- --....(((((((....((.((((...--........((((((...((....))...))))))..-.......(((....))))))).))..)))).))).....----- ( -21.10, z-score = -1.02, R) >droVir3.scaffold_12855 7649362 82 + 10161210 -----AGCCUCACACCUCCACACACUA------UCCCCUGCAGUAGUUG--CCGUUUUUAUGGUAAAUAUGUGGCGCAGGCGC------CA----GCCAUAAACA----- -----.((.(((((.........((((------(........)))))((--((((....))))))....))))).)).(((..------..----))).......----- ( -19.80, z-score = -0.50, R) >droMoj3.scaffold_6540 9139263 90 - 34148556 GAGUAAGCCCCACUCUGCCAAA---UU------UCCCCUACA-UAUUU----CAAGUUUAUAGCACAUUUGAGGCGCAAGCGU------CAAGGUGCUAUAAACAAACAU ((((.......)))).......---..------.........-.....----...(((((((((((.(((((.((....)).)------)))))))))))))))...... ( -30.00, z-score = -6.28, R) >consensus __GCUAACACCACCCAAGUAAAUCAUU__CGUAUUUCCCAUAAUUUGUAACCCCUUUUUAUGGAA_ACUCCAAGCGCAGGCGCGAUUAACAAGUGGCUGUAAACAA____ .........................................................................(((....)))........................... ( -3.91 = -3.82 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:27 2011