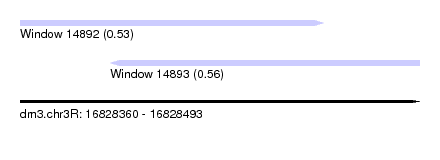
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,828,360 – 16,828,493 |
| Length | 133 |
| Max. P | 0.556336 |

| Location | 16,828,360 – 16,828,461 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.80 |
| Shannon entropy | 0.50739 |
| G+C content | 0.46691 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -9.52 |
| Energy contribution | -9.91 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
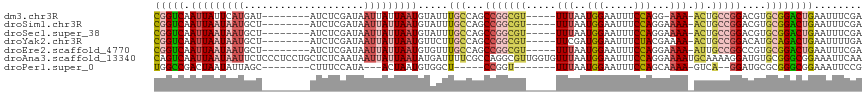
>dm3.chr3R 16828360 101 + 27905053 UCGCAAUCAAUCUGACGCUG---------AAGUUCCCUCUCGAAAUUCAGUCCGCACGUCCGGC-AGUUUUCC-UGGAAAUUCCAUUAAA--ACGCCGGCUGGCAAAUACAUUA--- ................((((---------((.(((......))).))))))..((.((.(((((-.(((((..-(((.....)))..)))--))))))).))))..........--- ( -27.80, z-score = -2.59, R) >droSim1.chr3R 22887435 102 - 27517382 UCGCAAUCAAUCUGACGCUG---------AAGUUCCCUCUCGAAAUUCAGUCCGCACGUCCGGC-AGUUUUUCCUGGAAAUUCCAUUAAA--ACGCCGGCUGGCAAAUACAUUA--- ................((((---------((.(((......))).))))))..((.((.(((((-.(((((...(((.....)))..)))--))))))).))))..........--- ( -27.00, z-score = -2.24, R) >droSec1.super_38 103607 102 - 400794 UCGCAAUCAAUCUGACGCUG---------AAGUUCCCUCUCGAAAUUCAGUCCGCACGUCCGGC-AGUUUUUCCUGGAAAUUCCAUUAAA--ACGCCGGCUGGCAAAUACAUUA--- ................((((---------((.(((......))).))))))..((.((.(((((-.(((((...(((.....)))..)))--))))))).))))..........--- ( -27.00, z-score = -2.24, R) >droYak2.chr3R 5626480 102 + 28832112 UCACAUUCGAUCUGACGCUG---------AAAUUCCCUCUCAAAAUUCAGUCUGCAUGUCCGGC-AGUUUUUCGUAGAAAUUCCAUCGAA--ACGCCGGCUGGCAAGAACAUUA--- ........(.(((...((((---------((.((........)).)))))).(((....(((((-.(((..(((..(.....)...))))--)))))))...)))))).)....--- ( -19.30, z-score = 0.38, R) >droEre2.scaffold_4770 12918912 102 - 17746568 UCACAAUCACUCUGACGCUG---------AAAUUCCCUCUCGAAAUUCAGUCCGCACGGCCGGC-AAUUUUUCCUGGAAAUUCCAUUAAA--ACGCCGGCUGGCAAACACAUUA--- ................((((---------((.(((......))).))))))..((.((((((((-...((((..(((.....)))..)))--).))))))))))..........--- ( -30.20, z-score = -4.20, R) >droAna3.scaffold_13340 8082575 117 + 23697760 UGGCAAUCAAUCUGGCGCCAGGACUCUGCAAUUUUCUUGUUGAAUUUCCGCCCGCACAUCCUUUUGCAUUUUCCUGGAAAUUCCAUUAAACACCAACGCCUGGCGAAAAUCAUAUUA ...............(((((((.....((((.....)))).(((((((((...(((........))).......)))))))))...............)))))))............ ( -23.60, z-score = -0.25, R) >droPer1.super_0 7857545 91 + 11822988 -GGCAAUCAAUCUG---CCAG-----CUCAAAUAUCUGGCGGAAUUUCCGCCCGCGCAUCCU---GACUUUUGCUGGAAAUUCCAUUAAA-ACCGGAGCCACAU------------- -(((.........(---((((-----.........)))))((((((((((....)(((....---......))).)))))))))......-......)))....------------- ( -27.10, z-score = -2.18, R) >consensus UCGCAAUCAAUCUGACGCUG_________AAAUUCCCUCUCGAAAUUCAGUCCGCACGUCCGGC_AGUUUUUCCUGGAAAUUCCAUUAAA__ACGCCGGCUGGCAAAUACAUUA___ .....................................................((.((.(((((.....(((..(((.....)))..)))....))))).))))............. ( -9.52 = -9.91 + 0.39)



| Location | 16,828,390 – 16,828,493 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.38 |
| Shannon entropy | 0.42766 |
| G+C content | 0.41027 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -14.13 |
| Energy contribution | -16.28 |
| Covariance contribution | 2.15 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16828390 103 - 27905053 CGGUCAAUUAUUCAUGAU--------AUCUCGAUAAUUAUUAAUGUAUUUGCCAGCCGGCGU-----UUUAAUGGAAUUUCCAGG-AAA-ACUGCCGGACGUGCGGACUGAAUUUCGA ((((((((((((...(..--------..)..)))))))...........((((..(((((((-----(((..(((.....)))..-)))-)).)))))..).))))))))........ ( -29.20, z-score = -1.88, R) >droSim1.chr3R 22887465 104 + 27517382 CGGUCAAUUAAUAAUGCU--------AUCUCGAUAAUUAUUAAUGUAUUUGCCAGCCGGCGU-----UUUAAUGGAAUUUCCAGGAAAA-ACUGCCGGACGUGCGGACUGAAUUUCGA (((((.(((((((((.(.--------.....)...))))))))).....((((..(((((((-----(((..(((.....)))...)))-)).)))))..).))))))))........ ( -29.70, z-score = -2.23, R) >droSec1.super_38 103637 104 + 400794 CGGUCAAUUAAUAAUGCU--------AUCUCGAUAAUUAUUAAUGUAUUUGCCAGCCGGCGU-----UUUAAUGGAAUUUCCAGGAAAA-ACUGCCGGACGUGCGGACUGAAUUUCGA (((((.(((((((((.(.--------.....)...))))))))).....((((..(((((((-----(((..(((.....)))...)))-)).)))))..).))))))))........ ( -29.70, z-score = -2.23, R) >droYak2.chr3R 5626510 104 - 28832112 CGGUCAAUUAAUAAUGCU--------AUCUCGAUAAUUAUUAAUGUUCUUGCCAGCCGGCGU-----UUCGAUGGAAUUUCUACGAAAA-ACUGCCGGACAUGCAGACUGAAUUUUGA (((((.(((((((((.(.--------.....)...))))))))).....(((...(((((((-----((((.(((.....)))))))).-..))))))....))))))))........ ( -29.00, z-score = -2.82, R) >droEre2.scaffold_4770 12918942 104 + 17746568 CGGUCAAUUAAUAAUGCU--------AUCUCGAUAAUUAUUAAUGUGUUUGCCAGCCGGCGU-----UUUAAUGGAAUUUCCAGGAAAA-AUUGCCGGCCGUGCGGACUGAAUUUCGA (((((.(((((((((.(.--------.....)...))))))))).....((((.((((((.(-----(((..(((.....)))..))))-...)))))).).))))))))........ ( -32.20, z-score = -2.44, R) >droAna3.scaffold_13340 8082614 118 - 23697760 CAGUCAAUUAAUAAUUCUCCCUCCUGCUCUCAAUAAUUAUUAAUAUGAUUUUCGCCAGGCGUUGGUGUUUAAUGGAAUUUCCAGGAAAAUGCAAAAGGAUGUGCGGGCGGAAAUUCAA ......((((((((((..................)))))))))).(((((((((((..(((((.((((((..(((.....)))...)))))).....)))))...)))))))).))). ( -24.77, z-score = 0.16, R) >droPer1.super_0 7857575 92 - 11822988 UGGCCGACUAAUAUUAGC--------CUUUCCAUA---ACUAAUGUGGCU-----CCGGU-------UUUAAUGGAAUUUCCAGCAAAA-GUCA--GGAUGCGCGGGCGGAAAUUCCG .(((((....(((((((.--------.........---.)))))))....-----.))))-------).....(((((((((.((....-((..--....))....))))))))))). ( -24.20, z-score = -0.36, R) >consensus CGGUCAAUUAAUAAUGCU________AUCUCGAUAAUUAUUAAUGUAUUUGCCAGCCGGCGU_____UUUAAUGGAAUUUCCAGGAAAA_ACUGCCGGACGUGCGGACUGAAUUUCGA (((((.(((((((((....................))))))))).....(((...((((((......(((..(((.....)))..)))....))))))....))))))))........ (-14.13 = -16.28 + 2.15)
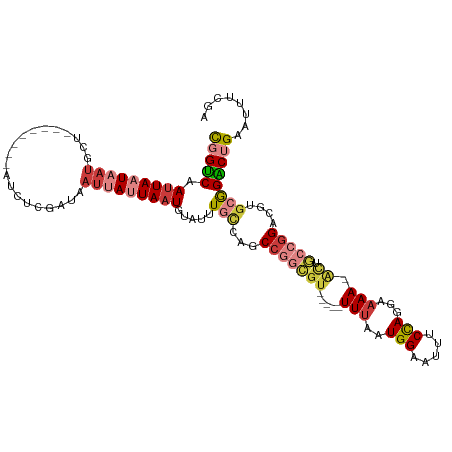
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:25 2011