| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,813,687 – 16,813,788 |
| Length | 101 |
| Max. P | 0.990379 |

| Location | 16,813,687 – 16,813,788 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.16 |
| Shannon entropy | 0.49171 |
| G+C content | 0.35099 |
| Mean single sequence MFE | -18.18 |
| Consensus MFE | -8.81 |
| Energy contribution | -10.56 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709532 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
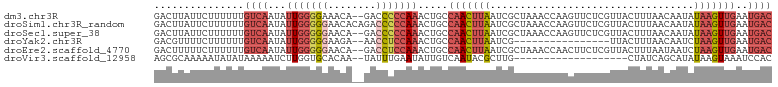
>dm3.chr3R 16813687 101 + 27905053 GACUUAUUCUUUUUUGUCAAUAUUGGGGAAACA--GACCCCCAAACUGCCAACUUAAUCGCUAAACCAAGUUCUCGUUACUUUAACAAUAUAAGUUGAAUGAC ...............((((...((((((.....--...)))))).....(((((((...(((......)))....((((...))))....)))))))..)))) ( -18.80, z-score = -1.83, R) >droSim1.chr3R_random 842746 103 + 1307089 GACUUAUUCUUUUUUGUCAAUAUUGGGGGAACACAGACCCCCAAACUGCCAACUUAAUCGCUAAACCAAGUUCUCGUUACUUUAACAAUAUAAGUUGAAUGAC ...............((((...(((((((........))))))).....(((((((...(((......)))....((((...))))....)))))))..)))) ( -21.70, z-score = -2.32, R) >droSec1.super_38 88575 101 - 400794 GACUUAUUCUUUUUUGUCAAUAUUGGGGGAACA--GACCCCCAAACUGCCAACUUAAUCGCUAAACCAAGUUCUCGUUACUUUAACAAUAUAAGUUGAAUGAC ...............((((...(((((((....--..))))))).....(((((((...(((......)))....((((...))))....)))))))..)))) ( -21.90, z-score = -2.39, R) >droYak2.chr3R 5605830 85 + 28832112 GACGUUUUCUUUUUUGUCAAUAUUGGGGGAAGA--AACCUCCAAACUGCCAACUUAAUCG----------------UUACUUUAACAAUCUAAGUUGAAUGAC ...............((((...(((((((....--..))))))).....(((((((((.(----------------(((...)))).)).)))))))..)))) ( -18.40, z-score = -1.88, R) >droEre2.scaffold_4770 12903494 101 - 17746568 GACUUUUUCUUUUUUGUCAAUAUUGGGGGAACA--GACCUCCAAACUGCCAACUUAAUCGCUAAACCAACUUCUCGUUACUUUAAUAAUCUAAGUUGAAUGAC ...............((((...(((((((....--..))))))).....(((((((.....((((..(((.....)))..))))......)))))))..)))) ( -18.10, z-score = -1.73, R) >droVir3.scaffold_12958 748438 82 + 3547706 AGCGCAAAAAUAUAUAAAAAUCUUGGUGCACAA--UAUUUGAAUAUUGUCAAUACGCUUG-------------------CUAUCAGCAUAUAAGUAAAUCCAC .((((...((.((......)).)).))))((((--(((....)))))))...(((...((-------------------(.....))).....)))....... ( -10.20, z-score = 0.42, R) >consensus GACUUAUUCUUUUUUGUCAAUAUUGGGGGAACA__GACCCCCAAACUGCCAACUUAAUCGCUAAACCAAGUUCUCGUUACUUUAACAAUAUAAGUUGAAUGAC ...............((((...(((((((........))))))).....(((((((..................................)))))))..)))) ( -8.81 = -10.56 + 1.75)
| Location | 16,813,687 – 16,813,788 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.16 |
| Shannon entropy | 0.49171 |
| G+C content | 0.35099 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -10.79 |
| Energy contribution | -14.33 |
| Covariance contribution | 3.55 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990379 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
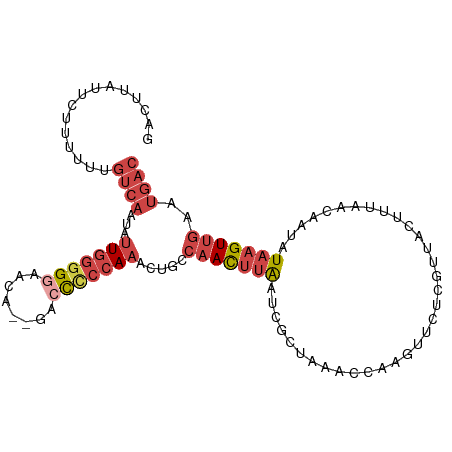
>dm3.chr3R 16813687 101 - 27905053 GUCAUUCAACUUAUAUUGUUAAAGUAACGAGAACUUGGUUUAGCGAUUAAGUUGGCAGUUUGGGGGUC--UGUUUCCCCAAUAUUGACAAAAAAGAAUAAGUC ((((.((((((((.(((((((((.(((.......))).)))))))))))))))))....(((((((..--....)))))))...))))............... ( -29.40, z-score = -3.72, R) >droSim1.chr3R_random 842746 103 - 1307089 GUCAUUCAACUUAUAUUGUUAAAGUAACGAGAACUUGGUUUAGCGAUUAAGUUGGCAGUUUGGGGGUCUGUGUUCCCCCAAUAUUGACAAAAAAGAAUAAGUC ((((.((((((((.(((((((((.(((.......))).)))))))))))))))))....(((((((........)))))))...))))............... ( -31.40, z-score = -3.93, R) >droSec1.super_38 88575 101 + 400794 GUCAUUCAACUUAUAUUGUUAAAGUAACGAGAACUUGGUUUAGCGAUUAAGUUGGCAGUUUGGGGGUC--UGUUCCCCCAAUAUUGACAAAAAAGAAUAAGUC ((((.((((((((.(((((((((.(((.......))).)))))))))))))))))....(((((((..--....)))))))...))))............... ( -31.60, z-score = -4.25, R) >droYak2.chr3R 5605830 85 - 28832112 GUCAUUCAACUUAGAUUGUUAAAGUAA----------------CGAUUAAGUUGGCAGUUUGGAGGUU--UCUUCCCCCAAUAUUGACAAAAAAGAAAACGUC ((((.(((((((.((((((((...)))----------------))))))))))))....((((.((..--....)).))))...))))............... ( -20.20, z-score = -2.50, R) >droEre2.scaffold_4770 12903494 101 + 17746568 GUCAUUCAACUUAGAUUAUUAAAGUAACGAGAAGUUGGUUUAGCGAUUAAGUUGGCAGUUUGGAGGUC--UGUUCCCCCAAUAUUGACAAAAAAGAAAAAGUC ((((.(((((((.(((..(((((.((((.....)))).)))))..))))))))))....((((.((..--....)).))))...))))............... ( -24.40, z-score = -2.68, R) >droVir3.scaffold_12958 748438 82 - 3547706 GUGGAUUUACUUAUAUGCUGAUAG-------------------CAAGCGUAUUGACAAUAUUCAAAUA--UUGUGCACCAAGAUUUUUAUAUAUUUUUGCGCU .(((((......(((((((.....-------------------..))))))).......)))))....--..(((((..(((((........)))))))))). ( -15.32, z-score = -0.60, R) >consensus GUCAUUCAACUUAUAUUGUUAAAGUAACGAGAACUUGGUUUAGCGAUUAAGUUGGCAGUUUGGAGGUC__UGUUCCCCCAAUAUUGACAAAAAAGAAUAAGUC ((((.((((((((.(((((((((.(((.......))).)))))))))))))))))....((((.((........)).))))...))))............... (-10.79 = -14.33 + 3.55)

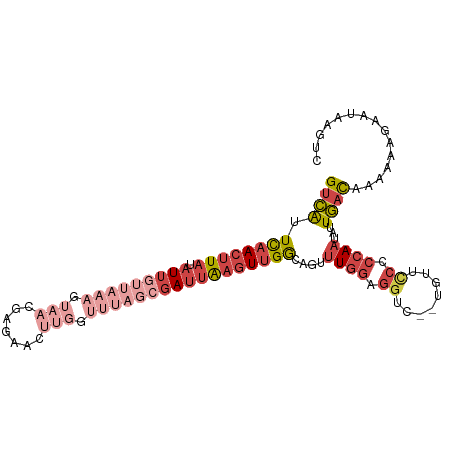
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:20 2011