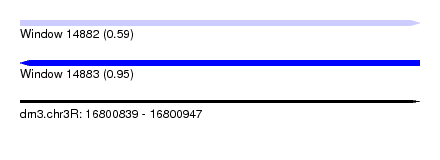
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,800,839 – 16,800,947 |
| Length | 108 |
| Max. P | 0.948338 |

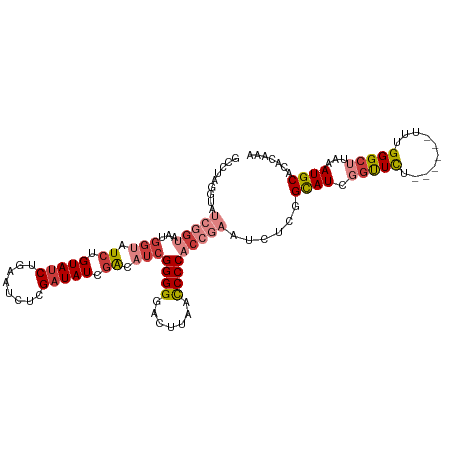
| Location | 16,800,839 – 16,800,947 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Shannon entropy | 0.36924 |
| G+C content | 0.46199 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -16.42 |
| Energy contribution | -18.63 |
| Covariance contribution | 2.21 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16800839 108 + 27905053 UUUGUGUGCAUUUAAGCCCAAA------AGAACCGAUGCCGAGAUUCGGUGGGAUUAAGUCCCCCGAUGUCGAUAUCGAGAUUCAGAUACAGAUACCAUUACCGAUACCUAGGC (((((((((......)).....------.(((((((((.(((.((.(((.(((((...)))))))))).))).))))).).)))..)))))))........((........)). ( -29.00, z-score = -2.49, R) >dp4.chr2 18158407 81 - 30794189 UUUGUGUGCAUUUAAGCCC---------AAAACAAAUACAGAGAUACAGAGGGGUUAGGUCCCCC----CCGAUAUCG--AUACCGAUACCUAAAU------------------ (((((((............---------.......)))))))((((..(.((((........)))----))..)))).--................------------------ ( -13.81, z-score = -0.18, R) >droAna3.scaffold_13340 8058408 100 + 23697760 UUUGUGUGCAUUUAAGCCCGAAAAACCAAAAACAAAUGCAGAGAUACGGAGGGGUUAAGACCCCCGAGAUCGAUAUCG--AUACCGUUACCGAUACCUAAGC------------ ..(.(.(((((((....................))))))).).)..(((.(((((....)))))((..((((....))--))..))...)))..........------------ ( -21.85, z-score = -1.85, R) >droEre2.scaffold_4770 12890400 108 - 17746568 UUUGUGUGCAUUUAAGCCCAAA------AGAAUCGAUGCCGAGAUUCGGUGGGGUUAAGUCCCCCGAUGUCGAUAUCGAGAUUCAGAUACAGAUACCAUUACCGAUACCUAGGC (((((((((......)).....------.(((((((((.(((.((.(((.((((.....))))))))).))).))))))..)))..)))))))........((........)). ( -28.40, z-score = -1.48, R) >droYak2.chr3R 5591613 108 + 28832112 UUUGUGUGCAUUUAAGCCCAAA------AGAACCGAUGCCGAGAUUCGGUGGGGUUAAGUCCCCCGAUGUCGAUAUCGAGAUUCAGAUACAGAUACCAUUACCGAUACCUAGGC (((((((((......)).....------.(((((((((.(((.((.(((.((((.....))))))))).))).))))).).)))..)))))))........((........)). ( -27.70, z-score = -1.41, R) >droSec1.super_38 75661 108 - 400794 UUUGUGUGCAUUUAAGCCCAAA------AGAACCGAUGCCGAGAUUCGGUGGGGUUAAGUCCCCCGAUGUCGAUAUCGAGAUUCAGAUACAGAUACCAUUACCGAUACCUAGGC (((((((((......)).....------.(((((((((.(((.((.(((.((((.....))))))))).))).))))).).)))..)))))))........((........)). ( -27.70, z-score = -1.41, R) >droSim1.chr3R 22866654 108 - 27517382 UUUGUGUGCAUUUAAGCCCAAA------AGAGCCGAUGCCGAGAUUCGGUGGGGUUAAGUCCCCCGAUGUCGAUAUCGAGAUUCAGAUACAGAUACCAUUACCGAUACCUAGGC (((((((((......)).....------.(((((((((.(((.((.(((.((((.....))))))))).))).))))).).)))..)))))))........((........)). ( -27.70, z-score = -1.22, R) >consensus UUUGUGUGCAUUUAAGCCCAAA______AGAACCGAUGCCGAGAUUCGGUGGGGUUAAGUCCCCCGAUGUCGAUAUCGAGAUUCAGAUACAGAUACCAUUACCGAUACCUAGGC (((((((((......))................(((((.(((.((.(((.((((.....))))))))).))).)))))........)))))))..................... (-16.42 = -18.63 + 2.21)

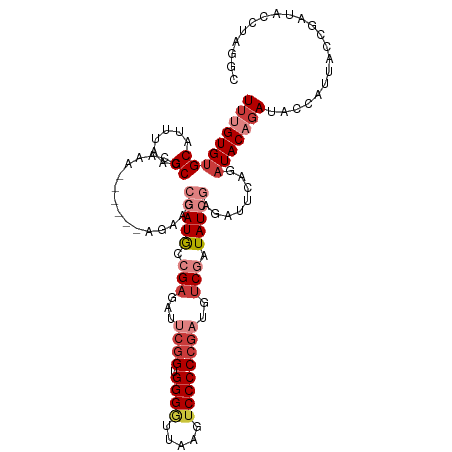
| Location | 16,800,839 – 16,800,947 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Shannon entropy | 0.36924 |
| G+C content | 0.46199 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -18.18 |
| Energy contribution | -19.96 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16800839 108 - 27905053 GCCUAGGUAUCGGUAAUGGUAUCUGUAUCUGAAUCUCGAUAUCGACAUCGGGGGACUUAAUCCCACCGAAUCUCGGCAUCGGUUCU------UUUGGGCUUAAAUGCACACAAA ((((((((((((....))))))).......((((..((((..(((..((((((((.....)))).))))...)))..)))))))).------..)))))............... ( -33.40, z-score = -2.43, R) >dp4.chr2 18158407 81 + 30794189 ------------------AUUUAGGUAUCGGUAU--CGAUAUCGG----GGGGGACCUAACCCCUCUGUAUCUCUGUAUUUGUUUU---------GGGCUUAAAUGCACACAAA ------------------.....(((((((....--)))))))((----(((((......))))))).......((((((((....---------.....))))))))...... ( -23.70, z-score = -1.66, R) >droAna3.scaffold_13340 8058408 100 - 23697760 ------------GCUUAGGUAUCGGUAACGGUAU--CGAUAUCGAUCUCGGGGGUCUUAACCCCUCCGUAUCUCUGCAUUUGUUUUUGGUUUUUCGGGCUUAAAUGCACACAAA ------------.....((((((((((....)))--)))))))(((..((((((........)))))).)))..((((((((..(((((....)))))..))))))))...... ( -31.40, z-score = -3.20, R) >droEre2.scaffold_4770 12890400 108 + 17746568 GCCUAGGUAUCGGUAAUGGUAUCUGUAUCUGAAUCUCGAUAUCGACAUCGGGGGACUUAACCCCACCGAAUCUCGGCAUCGAUUCU------UUUGGGCUUAAAUGCACACAAA (((.((((.(((((...(((.((.(((((........))))).)).)))((((.......))))))))))))).))).........------((((.((......))...)))) ( -33.30, z-score = -2.50, R) >droYak2.chr3R 5591613 108 - 28832112 GCCUAGGUAUCGGUAAUGGUAUCUGUAUCUGAAUCUCGAUAUCGACAUCGGGGGACUUAACCCCACCGAAUCUCGGCAUCGGUUCU------UUUGGGCUUAAAUGCACACAAA (((.((((.(((((...(((.((.(((((........))))).)).)))((((.......))))))))))))).))).........------((((.((......))...)))) ( -33.30, z-score = -2.16, R) >droSec1.super_38 75661 108 + 400794 GCCUAGGUAUCGGUAAUGGUAUCUGUAUCUGAAUCUCGAUAUCGACAUCGGGGGACUUAACCCCACCGAAUCUCGGCAUCGGUUCU------UUUGGGCUUAAAUGCACACAAA (((.((((.(((((...(((.((.(((((........))))).)).)))((((.......))))))))))))).))).........------((((.((......))...)))) ( -33.30, z-score = -2.16, R) >droSim1.chr3R 22866654 108 + 27517382 GCCUAGGUAUCGGUAAUGGUAUCUGUAUCUGAAUCUCGAUAUCGACAUCGGGGGACUUAACCCCACCGAAUCUCGGCAUCGGCUCU------UUUGGGCUUAAAUGCACACAAA (((.((((.(((((...(((.((.(((((........))))).)).)))((((.......))))))))))))).)))...(((((.------...))))).............. ( -35.60, z-score = -2.91, R) >consensus GCCUAGGUAUCGGUAAUGGUAUCUGUAUCUGAAUCUCGAUAUCGACAUCGGGGGACUUAACCCCACCGAAUCUCGGCAUCGGUUCU______UUUGGGCUUAAAUGCACACAAA .........(((((...(((.((.(((((........))))).)).)))((((.......)))))))))......((((.(((((..........)))))...))))....... (-18.18 = -19.96 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:16 2011