
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,762,527 – 16,762,623 |
| Length | 96 |
| Max. P | 0.628418 |

| Location | 16,762,527 – 16,762,623 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 65.08 |
| Shannon entropy | 0.59249 |
| G+C content | 0.51317 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -8.18 |
| Energy contribution | -8.57 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
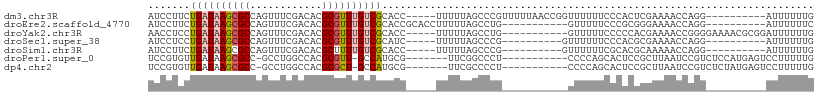
>dm3.chr3R 16762527 96 + 27905053 AUCCUUCUGAUAAGCGCCAGUUUCGACACGCGUUUGUCGCACC-----UUUUUAGCCCGUUUUUAACCGGUUUUUUCCCACUCGAAAACCAGG----------AUUUUUUG (((((...(((((((((..((....))..))))))))).....-----....................(((((((........))))))))))----------))...... ( -21.10, z-score = -2.36, R) >droEre2.scaffold_4770 12851739 90 - 17746568 AUCCUUCUGAUAAGCGCCAGUUUCGACACGCGUUUGUCGCACCGCACCUUUUUAGCCUG-----------GUUUUUCCCGCGGGAAAACCAGG----------AUUUUUUC ........(((((((((..((....))..))))))))).................((((-----------((((((((...))))))))))))----------........ ( -28.70, z-score = -3.09, R) >droYak2.chr3R 5566671 95 + 28832112 AACCUCCUGAUAAGCGCCAGUUUCGACACGCGUUUGUCGCACC-----UUUUUAGCCUG-----------GUUUUUCCCCCACGAAAACCGGGGAAAACGCGGAUUUUUUG ....(((.(((((((((..((....))..)))))))))(((((-----..........)-----------))((((((((..........)))))))).)))))....... ( -27.40, z-score = -1.68, R) >droSec1.super_38 36674 86 - 400794 AUCCUCCUGAUAAGCGCCAGUUUCGACACGCGUUUGUCGCAUC-----UUUUUAGCCCG----------GUUUUUUCCCACGCGAAAACCAGG----------AUUUUUUG ....(((((((((((((..((....))..))))))))).....-----..........(----------((((((........))))))))))----------)....... ( -21.20, z-score = -2.31, R) >droSim1.chr3R 22822358 86 - 27517382 AUCCUUCUGAUAAGCGCCAGUUUCGACACGCUUUUGUCGCACC-----UUUUUAGCCCG----------GUUUUUUCGCACGCAAAAACCAGG----------AUUUUUUG (((((.((((.(((.....((..(((((......))))).)))-----)).))))...(----------((((((.(....).))))))))))----------))...... ( -15.70, z-score = -0.40, R) >droPer1.super_0 7781881 91 + 11822988 UCCGUGUUGAUAAGCGCC-GCCUGGCCACGCGUU-GCCAUGCG-------UUCGGCCCU-----------CCCCAGCACUCCGCUUAAUCCGUCUCCAUGAGUCCUUUUUG ..((((..((((((((..-((..((((((((((.-...)))))-------)..))))..-----------.....))....))))).....)))..))))........... ( -21.30, z-score = -0.66, R) >dp4.chr2 18117509 91 - 30794189 UCCGUGUUGAUAAGCGCC-GCCUGGCCACGCGCU-GCCAUGCG-------UUCGCCCCU-----------CCCCAGCACUCCGCUUAAUCCGUCUCUAUGAGUCCUUUUUG ...((((((....(((.(-((.((((........-)))).)))-------..)))....-----------...))))))............(.(((...))).)....... ( -19.80, z-score = -0.78, R) >consensus AUCCUUCUGAUAAGCGCCAGUUUCGACACGCGUUUGUCGCACC_____UUUUUAGCCCG___________UUUUUUCCCACGCGAAAACCAGG__________AUUUUUUG .......((((((((((............))))))))))........................................................................ ( -8.18 = -8.57 + 0.39)
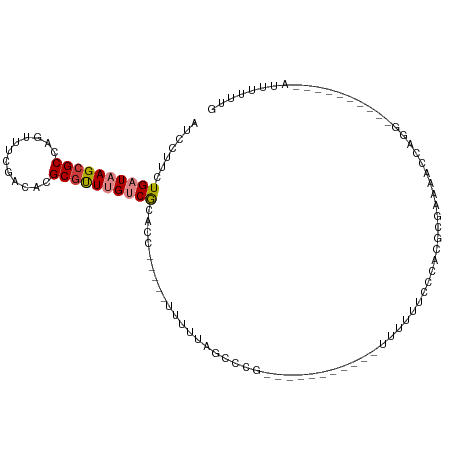

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:12 2011