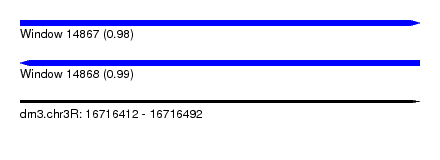
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,716,412 – 16,716,492 |
| Length | 80 |
| Max. P | 0.993881 |

| Location | 16,716,412 – 16,716,492 |
|---|---|
| Length | 80 |
| Sequences | 12 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 90.31 |
| Shannon entropy | 0.21949 |
| G+C content | 0.53328 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -25.29 |
| Energy contribution | -25.24 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
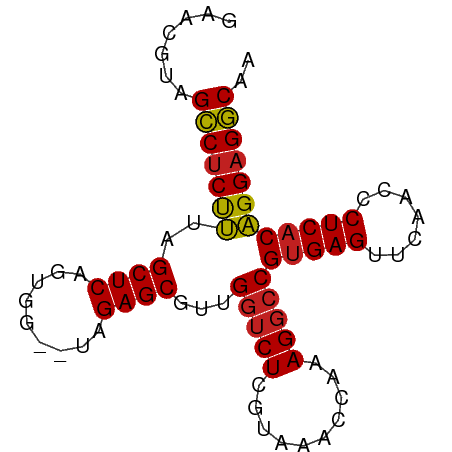
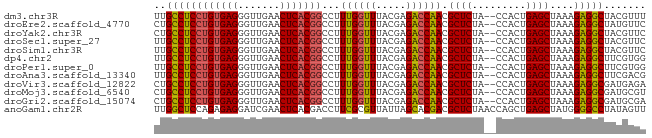
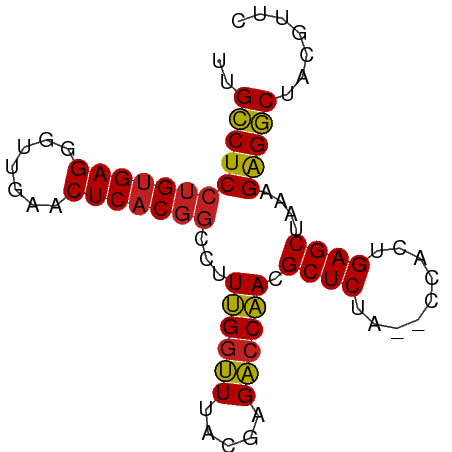
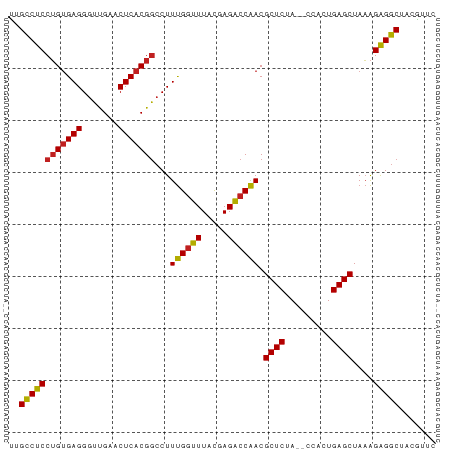
>dm3.chr3R 16716412 80 + 27905053 AAACGUAGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAA .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.80, z-score = -2.32, R) >droEre2.scaffold_4770 12805069 80 - 17746568 GAACAUAGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAG .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.80, z-score = -2.36, R) >droYak2.chr3R 5519529 80 + 28832112 GAACGUAGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAG .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.80, z-score = -2.05, R) >droSec1.super_27 781363 80 - 799419 GAACGUAGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAA .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.80, z-score = -2.05, R) >droSim1.chr3R 22773074 80 - 27517382 GAACGUAGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAA .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.80, z-score = -2.05, R) >dp4.chr2 18068658 80 - 30794189 CCACGAAGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAA .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.80, z-score = -2.04, R) >droPer1.super_0 7732505 80 + 11822988 CCACGAAGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAA .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.80, z-score = -2.04, R) >droAna3.scaffold_13340 7979136 80 + 23697760 CGUCGAAGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAA .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.80, z-score = -2.03, R) >droVir3.scaffold_12822 447802 80 + 4096053 UCUCAUCGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAG .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.90, z-score = -2.53, R) >droMoj3.scaffold_6540 33090018 80 + 34148556 ACGCAUCGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAG .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.90, z-score = -2.08, R) >droGri2.scaffold_15074 4392423 80 - 7742996 UCGCAUCGCCUCUUUAGCUCAGUGG--UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAG .......(((((((..((((.....--..))))...(((((..........)))))(((((.......)))))))))))).. ( -26.90, z-score = -2.24, R) >anoGam1.chr2R 13947725 82 - 62725911 AACUAUAGGCCCCAUAGCUCAGCUGGUUAGAGCGUCGUGCUAAUAACGCGAAGGUCGUGAGUUCGAUCCUCUCUGGAGCCAA .......(((.(((((((...))))...((((.((((.(((....((((....).))).))).)))).)))).))).))).. ( -25.90, z-score = -1.16, R) >consensus GAACGUAGCCUCUUUAGCUCAGUGG__UAGAGCGUUGGUCUCGUAAACCAAAGGCCGUGAGUUCAACCCUCACAGGAGGCAA .......(((((((..((((.........))))...(((((..........)))))(((((.......)))))))))))).. (-25.29 = -25.24 + -0.05)

| Location | 16,716,412 – 16,716,492 |
|---|---|
| Length | 80 |
| Sequences | 12 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 90.31 |
| Shannon entropy | 0.21949 |
| G+C content | 0.53328 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -27.17 |
| Energy contribution | -26.97 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.993881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16716412 80 - 27905053 UUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCUACGUUU ..((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....)))))....... ( -28.40, z-score = -2.76, R) >droEre2.scaffold_4770 12805069 80 + 17746568 CUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCUAUGUUC ..((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....)))))....... ( -28.40, z-score = -2.72, R) >droYak2.chr3R 5519529 80 - 28832112 CUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCUACGUUC ..((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....)))))....... ( -28.40, z-score = -2.54, R) >droSec1.super_27 781363 80 + 799419 UUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCUACGUUC ..((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....)))))....... ( -28.40, z-score = -2.63, R) >droSim1.chr3R 22773074 80 + 27517382 UUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCUACGUUC ..((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....)))))....... ( -28.40, z-score = -2.63, R) >dp4.chr2 18068658 80 + 30794189 UUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCUUCGUGG ..((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....)))))....... ( -28.40, z-score = -2.13, R) >droPer1.super_0 7732505 80 - 11822988 UUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCUUCGUGG ..((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....)))))....... ( -28.40, z-score = -2.13, R) >droAna3.scaffold_13340 7979136 80 - 23697760 UUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCUUCGACG ..((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....)))))....... ( -28.40, z-score = -2.74, R) >droVir3.scaffold_12822 447802 80 - 4096053 CUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCGAUGAGA .(((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....))))))...... ( -29.00, z-score = -2.78, R) >droMoj3.scaffold_6540 33090018 80 - 34148556 CUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCGAUGCGU .(((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....))))))...... ( -29.00, z-score = -2.46, R) >droGri2.scaffold_15074 4392423 80 + 7742996 CUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA--CCACUGAGCUAAAGAGGCGAUGCGA .(((((((((((((.......)))))))...((((((.....)))))).((((..--.....))))....))))))...... ( -29.00, z-score = -2.65, R) >anoGam1.chr2R 13947725 82 + 62725911 UUGGCUCCAGAGAGGAUCGAACUCACGACCUUCGCGUUAUUAGCACGACGCUCUAACCAGCUGAGCUAUGGGGCCUAUAGUU ..(((((((..((((.(((......))).))))((.......)).....((((.........))))..)))))))....... ( -25.60, z-score = -1.38, R) >consensus UUGCCUCCUGUGAGGGUUGAACUCACGGCCUUUGGUUUACGAGACCAACGCUCUA__CCACUGAGCUAAAGAGGCUACGUUC ..((((((((((((.......)))))))...((((((.....)))))).((((.........))))....)))))....... (-27.17 = -26.97 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:04 2011