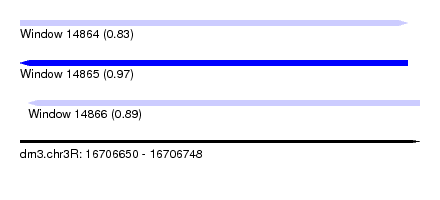
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,706,650 – 16,706,748 |
| Length | 98 |
| Max. P | 0.973628 |

| Location | 16,706,650 – 16,706,745 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Shannon entropy | 0.22199 |
| G+C content | 0.47046 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -26.26 |
| Energy contribution | -27.14 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16706650 95 + 27905053 -----------UGCUUUCUGUUUUGACAUUCAUUUGCCAAAGUC--CACAGAAAGCGUUUGCAAUUUGCCGAGGCAUCAUUGCGCAACUGGGGAGCCGGCAGUGAACU -----------.(((((((((((((.((......)).))))...--.)))))))))(((((((((.(((....)))..)))))((..((((....))))..)))))). ( -31.00, z-score = -1.31, R) >droEre2.scaffold_4770 12795195 100 - 17746568 UGCCAUUUGAAUACUUUCUGUUUUGACAUUCAUUUGCGAAAGUC--CACAGAAAGCGUUUGCAAUUUGCCGAGGCAU---UGCGCAACUGGGGAGCCAGC---GAACU (((.....(((..((((((((...(((.(((......))).)))--.))))))))..)))((((..(((....))))---)))))).((((....)))).---..... ( -30.40, z-score = -1.59, R) >droYak2.chr3R 5508487 100 + 28832112 UGCCAUUUGAAUACUUUCUGUUUUGACAUUCAUUUGCGAAAGUC--CACAGAAAGCGUUUGCAAUUUGCCGACGCAU---UGCGCAACUGGGAAGCAAGC---GAACU .............((((((((...(((.(((......))).)))--.))))))))(((((((...((.(((..((..---...))...))).))))))))---).... ( -25.50, z-score = -0.51, R) >droSec1.super_27 771431 94 - 799419 -----------UGCUUUCUGUUUUGACAUUCAUUUGGGAAAGUCCCCACAGAAAGCGUUUGCAAUUUGCCGAGGCAUCAUUGCGCAACUGGGGAGUCAGC---GAACU -----------.(((((((((...(((.(((......))).)))...)))))))))((((((.((((.(((..((........))...))).))))..))---)))). ( -32.10, z-score = -2.08, R) >droSim1.chr3R 22763179 94 - 27517382 -----------UGCUUUCUGUUUUGACAUUCAUUUGGGAAAGUCCCCACAGAAAGCGUUUGCAAUUUGCCGAGGCAUCAUUGCGCAACUGGGGAGCCAGC---GAACU -----------.(((((((((...(((.(((......))).)))...)))))))))(((((((((.(((....)))..)))))....((((....)))).---)))). ( -34.20, z-score = -2.52, R) >consensus ___________UGCUUUCUGUUUUGACAUUCAUUUGCGAAAGUC__CACAGAAAGCGUUUGCAAUUUGCCGAGGCAUCAUUGCGCAACUGGGGAGCCAGC___GAACU ............(((((((((...(((.(((......))).)))...)))))))))((((((((..(((....)))...))))....((((....))))....)))). (-26.26 = -27.14 + 0.88)

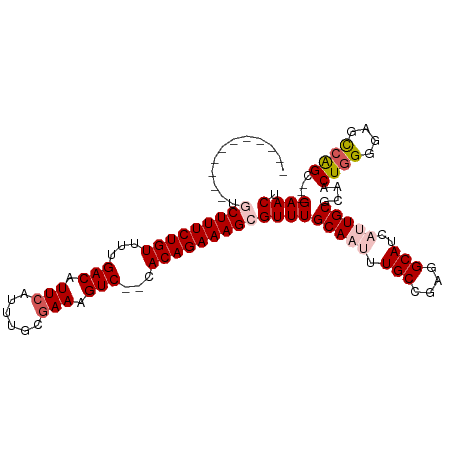
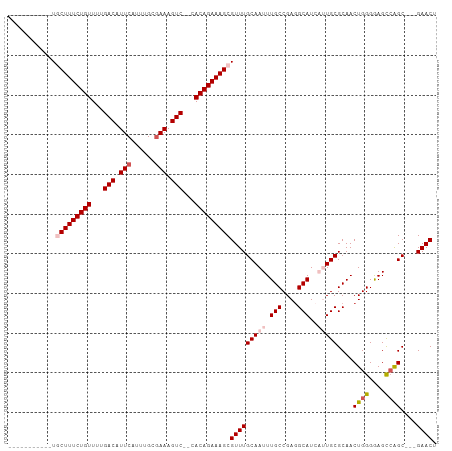
| Location | 16,706,650 – 16,706,745 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.88 |
| Shannon entropy | 0.22199 |
| G+C content | 0.47046 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16706650 95 - 27905053 AGUUCACUGCCGGCUCCCCAGUUGCGCAAUGAUGCCUCGGCAAAUUGCAAACGCUUUCUGUG--GACUUUGGCAAAUGAAUGUCAAAACAGAAAGCA----------- ...(((.((((((((....))))).))).)))(((....)))..........(((((((((.--...(((((((......)))))))))))))))).----------- ( -30.90, z-score = -1.78, R) >droEre2.scaffold_4770 12795195 100 + 17746568 AGUUC---GCUGGCUCCCCAGUUGCGCA---AUGCCUCGGCAAAUUGCAAACGCUUUCUGUG--GACUUUCGCAAAUGAAUGUCAAAACAGAAAGUAUUCAAAUGGCA .....---(((((....)))))((((((---((((....)))..))))....(((((((((.--(((.((((....)))).)))...))))))))).........))) ( -31.90, z-score = -2.34, R) >droYak2.chr3R 5508487 100 - 28832112 AGUUC---GCUUGCUUCCCAGUUGCGCA---AUGCGUCGGCAAAUUGCAAACGCUUUCUGUG--GACUUUCGCAAAUGAAUGUCAAAACAGAAAGUAUUCAAAUGGCA .....---((((((......(((((((.---..))).)))).....))))..(((((((((.--(((.((((....)))).)))...))))))))).........)). ( -27.70, z-score = -0.94, R) >droSec1.super_27 771431 94 + 799419 AGUUC---GCUGACUCCCCAGUUGCGCAAUGAUGCCUCGGCAAAUUGCAAACGCUUUCUGUGGGGACUUUCCCAAAUGAAUGUCAAAACAGAAAGCA----------- .((..---((((......)))).))(((((..(((....))).)))))....(((((((((...(((.(((......))).)))...))))))))).----------- ( -33.60, z-score = -3.43, R) >droSim1.chr3R 22763179 94 + 27517382 AGUUC---GCUGGCUCCCCAGUUGCGCAAUGAUGCCUCGGCAAAUUGCAAACGCUUUCUGUGGGGACUUUCCCAAAUGAAUGUCAAAACAGAAAGCA----------- .((..---(((((....))))).))(((((..(((....))).)))))....(((((((((...(((.(((......))).)))...))))))))).----------- ( -35.90, z-score = -3.49, R) >consensus AGUUC___GCUGGCUCCCCAGUUGCGCAAUGAUGCCUCGGCAAAUUGCAAACGCUUUCUGUG__GACUUUCGCAAAUGAAUGUCAAAACAGAAAGCA___________ .(((....((.((((....))))))(((((..(((....))).))))).)))(((((((((...(((.(((......))).)))...)))))))))............ (-26.72 = -27.52 + 0.80)

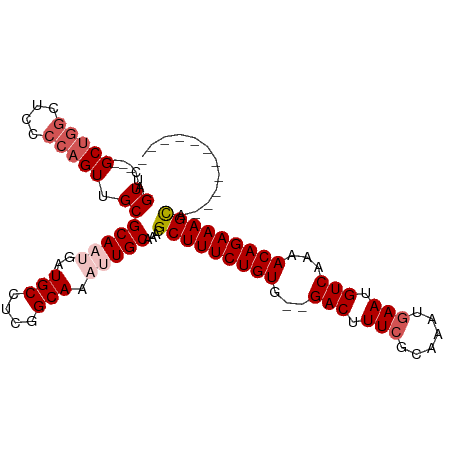
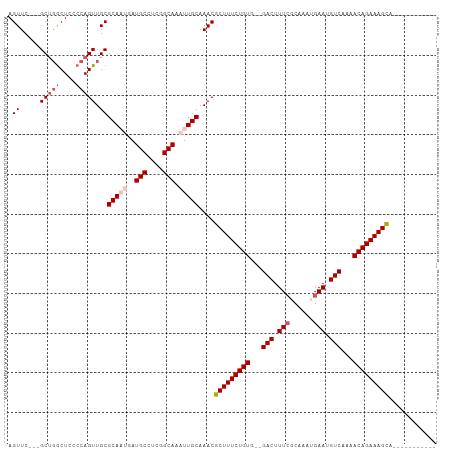
| Location | 16,706,652 – 16,706,748 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Shannon entropy | 0.20401 |
| G+C content | 0.47371 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -22.90 |
| Energy contribution | -23.94 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16706652 96 - 27905053 CCAAGUUCACUGCCGGCUCCCCAGUUGCGCAAUGAUGCCUCGGCAAAUUGCAAACGCUUUCUGU--GGACUUUGGCAAAUGAAUGUCAAAACAGAAAG---------- ....(((((.((((((.(((.(((..(((((((..(((....))).)))))....))...))).--)))..))))))..)))))..............---------- ( -26.60, z-score = -0.96, R) >droEre2.scaffold_4770 12795198 100 + 17746568 CCAAGUUC---GCUGGCUCCCCAGUUGCGCA---AUGCCUCGGCAAAUUGCAAACGCUUUCUGU--GGACUUUCGCAAAUGAAUGUCAAAACAGAAAGUAUUCAAAUG ....((..---(((((....))))).))(((---((((....)))..))))....(((((((((--.(((.((((....)))).)))...)))))))))......... ( -30.90, z-score = -3.04, R) >droYak2.chr3R 5508490 100 - 28832112 CCAAGUUC---GCUUGCUUCCCAGUUGCGCA---AUGCGUCGGCAAAUUGCAAACGCUUUCUGU--GGACUUUCGCAAAUGAAUGUCAAAACAGAAAGUAUUCAAAUG .((((...---.)))).......(((..(((---((((....)))..)))).)))(((((((((--.(((.((((....)))).)))...)))))))))......... ( -27.30, z-score = -1.72, R) >droSec1.super_27 771433 95 + 799419 CCAAGUUC---GCUGACUCCCCAGUUGCGCAAUGAUGCCUCGGCAAAUUGCAAACGCUUUCUGUGGGGACUUUCCCAAAUGAAUGUCAAAACAGAAAG---------- ....((..---((((......)))).))(((((..(((....))).))))).....((((((((...(((.(((......))).)))...))))))))---------- ( -28.40, z-score = -2.29, R) >droSim1.chr3R 22763181 95 + 27517382 CCAAGUUC---GCUGGCUCCCCAGUUGCGCAAUGAUGCCUCGGCAAAUUGCAAACGCUUUCUGUGGGGACUUUCCCAAAUGAAUGUCAAAACAGAAAG---------- ....((..---(((((....))))).))(((((..(((....))).))))).....((((((((...(((.(((......))).)))...))))))))---------- ( -30.70, z-score = -2.39, R) >consensus CCAAGUUC___GCUGGCUCCCCAGUUGCGCAAUGAUGCCUCGGCAAAUUGCAAACGCUUUCUGU__GGACUUUCGCAAAUGAAUGUCAAAACAGAAAG__________ ....(((....((.((((....))))))(((((..(((....))).))))).))).((((((((...(((.(((......))).)))...)))))))).......... (-22.90 = -23.94 + 1.04)

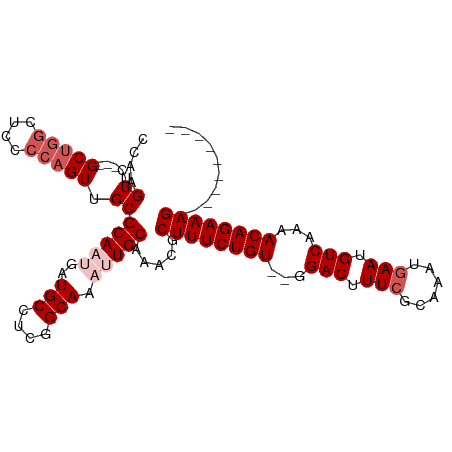
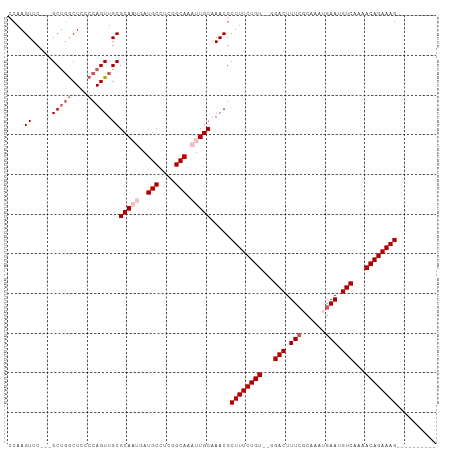
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:31:02 2011