| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,648,400 – 16,648,494 |
| Length | 94 |
| Max. P | 0.999973 |

| Location | 16,648,400 – 16,648,494 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 56.25 |
| Shannon entropy | 0.84695 |
| G+C content | 0.38323 |
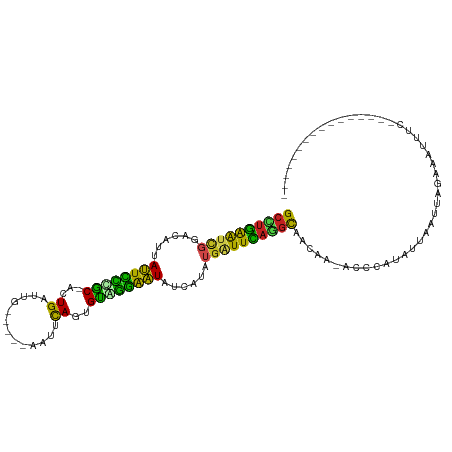
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -20.15 |
| Energy contribution | -17.84 |
| Covariance contribution | -2.30 |
| Combinations/Pair | 2.00 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.47 |
| SVM RNA-class probability | 0.999973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
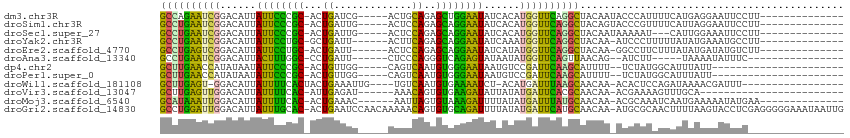
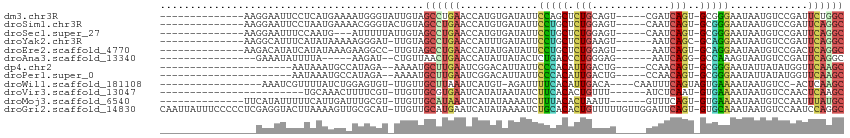
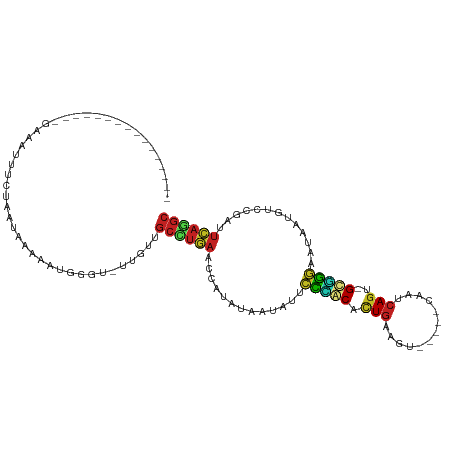
>dm3.chr3R 16648400 94 + 27905053 GCCAGAAUCGGACAUUAUUCCCGC-ACUGAUCG-----ACUGCAGAGCUGGAAUAUCACAUGGUUCAGGCUACAAUACCCAUUUUCAUGAGGAAUUCCUU-------------- (((.((((((.....((((((.((-.(((....-----....))).)).)))))).....)))))).)))..................((((....))))-------------- ( -26.70, z-score = -2.42, R) >droSim1.chr3R 22704677 94 - 27517382 GCCUGAAUCGGACAUUAUUCCCGC-ACUGAUUG-----ACUCCAGAGCAGGAAUAUCACAUGGUUCAGGCUACAGUACCCGUUUUCAUUAGGAAUUCCUU-------------- ((((((((((.....((((((.((-.(((....-----....))).)).)))))).....))))))))))...................(((....))).-------------- ( -31.00, z-score = -3.96, R) >droSec1.super_27 711946 91 - 799419 GCCUGAAUCGGACAUUAUUCCCGC-ACUGAUUG-----ACUCCAGAGCAGGAAUAUCACAUGGUUCAGGCUACAAUAAAAAU---CAUUGGAAAUUCCUU-------------- ((((((((((.....((((((.((-.(((....-----....))).)).)))))).....))))))))))............---....((.....))..-------------- ( -30.30, z-score = -4.47, R) >droYak2.chr3R 5444282 92 + 28832112 GCCUGAAUCGGACAUUAUUCCUGC-GCUGAUU------ACUUCAGAGCAGGAAUAUCAAAUGGUUCAGGCUACAA-AUCCCUUUUUAUAUGAAAUGCCUU-------------- ((((((((((.....(((((((((-.((((..------...)))).))))))))).....)))))))))).....-........................-------------- ( -36.40, z-score = -6.41, R) >droEre2.scaffold_4770 12731725 92 - 17746568 GCCUGAGUCGGACAUUAUUCCUGC-ACUGAUU------ACUCCAGAGCAGGAAUAUCAUAUGGUUCAGGCUACAA-GGCCUUCUUUAUAUGAUAUGUCUU-------------- ((((((..((.....(((((((((-.(((...------....))).))))))))).....))..))))))...((-(((..((.......))...)))))-------------- ( -34.90, z-score = -4.66, R) >droAna3.scaffold_13340 7915100 84 + 23697760 GCCUGAAUCGGACAUUACUUUGGC-CCUGAUU------CUCCCAGGGUCAGAGUAUAAUAUGGUUCAGUUAACAG--AUCUU-----UAAAAUAUUUC---------------- ..((((((((.....(((((((((-((((...------....))))))))))))).....)))))))).......--.....-----...........---------------- ( -30.60, z-score = -5.05, R) >dp4.chr2 18005968 84 - 30794189 GCUUGAACCAUAUAAUAUUCCCGC-ACUGUUGG-----CAGUCAAUGUGGGAAUAAUGUCCGAUUCAAGCAUUUU--UCUAUGGCAUUUAUU---------------------- (((((((.(..(((.(((((((((-((((....-----))))....))))))))).)))..).))))))).....--...............---------------------- ( -26.10, z-score = -3.47, R) >droPer1.super_0 7670350 84 + 11822988 GCUUGAACCAUAUAAUAUUCCCGC-ACUGUUGG-----CAGUCAAUGUGGGAAUAAUGUCCGAUUCAAGCAUUUU--UCUAUGGCAUUUAUU---------------------- (((((((.(..(((.(((((((((-((((....-----))))....))))))))).)))..).))))))).....--...............---------------------- ( -26.10, z-score = -3.47, R) >droWil1.scaffold_181108 2491663 90 - 4707319 GCUUGAGU-GGACAUUAUUUUCACUACUGAAAUUG----UGUCAAUGUGAAAAUCU-ACAUGAUUUAAGCAACAA-ACACUCCAGAUAAAACGAUUU----------------- .((.((((-((((((...(((((....)))))..)----))))..(((..(((((.-....)))))..)))....-.))))).))............----------------- ( -21.90, z-score = -3.29, R) >droVir3.scaffold_13047 12060386 82 + 19223366 GCUUGAGUUGGACAUUAUUUUCAC-AUUGAGAU------AAACAGUGUGAAGAUAUUAUAUGAUUCACGCAACAA-ACGAAAAGUUUGCA------------------------ ((.(((((((.....(((((((((-((((....------...))))))))))))).....))))))).))..(((-((.....)))))..------------------------ ( -26.30, z-score = -5.21, R) >droMoj3.scaffold_6540 4428208 92 - 34148556 GCAUAAAUUGGACAUUAUUUUCAC-ACUGAAAC------AAUUAGUGUAAAGAUUUUAUAUGAUUUAUGCAACAA-ACGCAAAUCAAUGAAAAAUAUGAA-------------- ((((((((((......(((((.((-(((((...------..))))))).)))))......)))))))))).....-........................-------------- ( -19.00, z-score = -3.12, R) >droGri2.scaffold_14830 5115794 112 - 6267026 GCCUGGAUUGGACAUUAUUUGCAC-ACUGAAUCCAACAAAAACAGUGUGCAGAUUUUAUAUGAUUCAUGCAACAA-AUGCGCAACUUUUAAGUACCUCGAGGGGGAAAUAAUUG ((.(((((((......((((((((-((((.............))))))))))))......))))))).)).....-..................(((....))).......... ( -27.62, z-score = -2.06, R) >consensus GCCUGAAUCGGACAUUAUUCCCGC_ACUGAUUG_____AAUUCAGUGUAGGAAUAUCAUAUGAUUCAGGCAACAA_ACCCAUAUUAAUUAGAAAUUUC________________ ((((((((((......((((((((...((.............))..))))))))......))))))))))............................................ (-20.15 = -17.84 + -2.30)
| Location | 16,648,400 – 16,648,494 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 56.25 |
| Shannon entropy | 0.84695 |
| G+C content | 0.38323 |
| Mean single sequence MFE | -23.61 |
| Consensus MFE | -8.24 |
| Energy contribution | -7.09 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.91 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.996236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16648400 94 - 27905053 --------------AAGGAAUUCCUCAUGAAAAUGGGUAUUGUAGCCUGAACCAUGUGAUAUUCCAGCUCUGCAGU-----CGAUCAGU-GCGGGAAUAAUGUCCGAUUCUGGC --------------..((((((.(.((((.....((((......))))....)))).).((((((.((.(((....-----....))).-)).))))))......))))))... ( -22.00, z-score = 0.37, R) >droSim1.chr3R 22704677 94 + 27517382 --------------AAGGAAUUCCUAAUGAAAACGGGUACUGUAGCCUGAACCAUGUGAUAUUCCUGCUCUGGAGU-----CAAUCAGU-GCGGGAAUAAUGUCCGAUUCAGGC --------------.(((....))).......((((...)))).(((((((........(((((((((.((((...-----...)))).-)))))))))........))))))) ( -29.19, z-score = -2.23, R) >droSec1.super_27 711946 91 + 799419 --------------AAGGAAUUUCCAAUG---AUUUUUAUUGUAGCCUGAACCAUGUGAUAUUCCUGCUCUGGAGU-----CAAUCAGU-GCGGGAAUAAUGUCCGAUUCAGGC --------------..((.....))....---............(((((((........(((((((((.((((...-----...)))).-)))))))))........))))))) ( -27.99, z-score = -2.46, R) >droYak2.chr3R 5444282 92 - 28832112 --------------AAGGCAUUUCAUAUAAAAAGGGAU-UUGUAGCCUGAACCAUUUGAUAUUCCUGCUCUGAAGU------AAUCAGC-GCAGGAAUAAUGUCCGAUUCAGGC --------------...(((..((...........)).-.))).(((((((........(((((((((.((((...------..)))).-)))))))))........))))))) ( -29.69, z-score = -3.42, R) >droEre2.scaffold_4770 12731725 92 + 17746568 --------------AAGACAUAUCAUAUAAAGAAGGCC-UUGUAGCCUGAACCAUAUGAUAUUCCUGCUCUGGAGU------AAUCAGU-GCAGGAAUAAUGUCCGACUCAGGC --------------..(((((.((((((.....((((.-.....)))).....))))))(((((((((.((((...------..)))).-)))))))))))))).......... ( -32.10, z-score = -4.10, R) >droAna3.scaffold_13340 7915100 84 - 23697760 ----------------GAAAUAUUUUA-----AAGAU--CUGUUAACUGAACCAUAUUAUACUCUGACCCUGGGAG------AAUCAGG-GCCAAAGUAAUGUCCGAUUCAGGC ----------------...........-----.....--.......(((((........((((.((.((((((...------..)))))-).)).))))........))))).. ( -18.09, z-score = -0.81, R) >dp4.chr2 18005968 84 + 30794189 ----------------------AAUAAAUGCCAUAGA--AAAAUGCUUGAAUCGGACAUUAUUCCCACAUUGACUG-----CCAACAGU-GCGGGAAUAUUAUAUGGUUCAAGC ----------------------...............--.....((((((((((.....(((((((.(((((....-----....))))-).))))))).....)))))))))) ( -28.40, z-score = -4.79, R) >droPer1.super_0 7670350 84 - 11822988 ----------------------AAUAAAUGCCAUAGA--AAAAUGCUUGAAUCGGACAUUAUUCCCACAUUGACUG-----CCAACAGU-GCGGGAAUAUUAUAUGGUUCAAGC ----------------------...............--.....((((((((((.....(((((((.(((((....-----....))))-).))))))).....)))))))))) ( -28.40, z-score = -4.79, R) >droWil1.scaffold_181108 2491663 90 + 4707319 -----------------AAAUCGUUUUAUCUGGAGUGU-UUGUUGCUUAAAUCAUGU-AGAUUUUCACAUUGACA----CAAUUUCAGUAGUGAAAAUAAUGUCC-ACUCAAGC -----------------...............(((((.-.((((((.........))-..(((((((((((((..----.....))))).))))))))))))..)-)))).... ( -17.20, z-score = -0.87, R) >droVir3.scaffold_13047 12060386 82 - 19223366 ------------------------UGCAAACUUUUCGU-UUGUUGCGUGAAUCAUAUAAUAUCUUCACACUGUUU------AUCUCAAU-GUGAAAAUAAUGUCCAACUCAAGC ------------------------.((((((.....))-)))).((.(((.....(((.(((.((((((.((...------....)).)-))))).))).))).....))).)) ( -14.20, z-score = -1.73, R) >droMoj3.scaffold_6540 4428208 92 + 34148556 --------------UUCAUAUUUUUCAUUGAUUUGCGU-UUGUUGCAUAAAUCAUAUAAAAUCUUUACACUAAUU------GUUUCAGU-GUGAAAAUAAUGUCCAAUUUAUGC --------------..((((..((.((((((((((((.-....))))..))))..........((((((((....------.....)))-)))))...))))...))..)))). ( -13.30, z-score = -0.49, R) >droGri2.scaffold_14830 5115794 112 + 6267026 CAAUUAUUUCCCCCUCGAGGUACUUAAAAGUUGCGCAU-UUGUUGCAUGAAUCAUAUAAAAUCUGCACACUGUUUUUGUUGGAUUCAGU-GUGCAAAUAAUGUCCAAUCCAGGC ............(((.((((.((...........(((.-....))).................(((((((((..((.....))..))))-)))))......))))..)).))). ( -22.80, z-score = -0.46, R) >consensus ________________GAAAUUUCUAAUAAAAAUGGGU_UUGUUGCCUGAACCAUAUAAUAUUCCCACACUGAAGU_____CAAUCAGU_GCGGGAAUAAUGUCCGAUUCAGGC ............................................((((((.............(((((......................))))).............)))))) ( -8.24 = -7.09 + -1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:56 2011