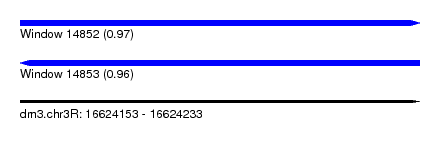
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,624,153 – 16,624,233 |
| Length | 80 |
| Max. P | 0.965046 |

| Location | 16,624,153 – 16,624,233 |
|---|---|
| Length | 80 |
| Sequences | 7 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 65.47 |
| Shannon entropy | 0.65469 |
| G+C content | 0.45962 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -10.90 |
| Energy contribution | -11.69 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.965046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
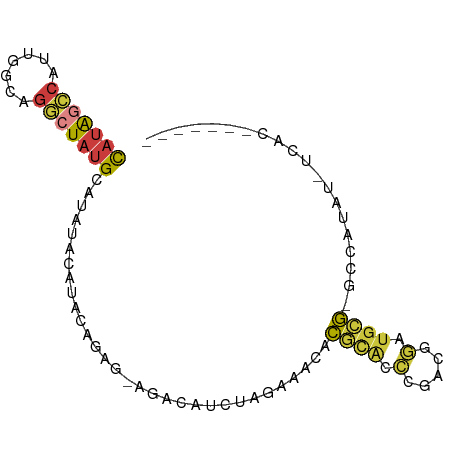
>dm3.chr3R 16624153 80 + 27905053 -------GUGA-AUAUGGC-CGCAUCCGUCGGAUGCGUGUUACUAGAUGUCUUCUCUGUAUGUAUAUGCAUAGCCUGCCAAUGGCUAUG -------....-(((((((-(((((((...))))))).))(((.(((.....)))..)))..))))).(((((((.......))))))) ( -23.20, z-score = -0.95, R) >droSim1.chr3R 22679596 79 - 27517382 -------GUGA-AUAUGGC-CGCAUCCGUCGGAUGCGUGUUUCUAGAUGUCU-CUCUGUAUGUAUAUGCAUAGCCUGCCAAUGGCUAUG -------(((.-(((((((-(((((((...))))))).))...((((.....-.))))))))).))).(((((((.......))))))) ( -22.50, z-score = -0.96, R) >droSec1.super_27 687535 79 - 799419 -------GUGA-AUAUGGC-CGCAUCCGUCGGAUGCGUGUUUCUAGAUGUCU-CUCUGUAUGUAUAUGCAUAGCCUGCCAAUGGCUAUG -------(((.-(((((((-(((((((...))))))).))...((((.....-.))))))))).))).(((((((.......))))))) ( -22.50, z-score = -0.96, R) >droYak2.chr3R 5419346 78 + 28832112 -------GUGA-AUAUGGC-CGCAUCCGUCGGGUGCGUGUU-CUAGAUGUCU-CUCUGUAUGUAUAUGCAUAGCCCCCCGAGGGCUAUG -------(.((-.((..((-(((((((...))))))).)).-.))....)).-)..............((((((((.....)))))))) ( -24.60, z-score = -0.93, R) >droEre2.scaffold_4770 12706761 78 - 17746568 -------GUGA-AUAUGUC-CGCAUCCAACGGGUGCGUGUU-CUAGAUGUCU-CUCUGUAUGUAUAUGCAUAGCCGCCCAAGGGCUAUG -------..((-(((....-(((((((...)))))))))))-)((((.....-.))))..........(((((((.......))))))) ( -21.00, z-score = -0.39, R) >droWil1.scaffold_181108 2466549 88 - 4707319 GUGACAUAUGACACACGACAUACAAUCAAUCAGCAUAUGUGUUUAUUAACUUGGCCUAUAGGCCAAAGCAUAAACA-CAUAUGUAUGUA (((........)))..................((((((((((((((....((((((....))))))...)))))))-)))))))..... ( -27.90, z-score = -4.46, R) >droMoj3.scaffold_6540 29695606 77 + 34148556 -------GUGA-AC-CAAU-CGAUUAUUCUCAUCAAGUAUAUCAAGA-AUAU-CUGAGUGUAUACAUACUUAACGAGCCCCCGCACAUG -------(((.-..-...(-((..((((((..............)))-))).-.(((((((....))))))).))).......)))... ( -11.56, z-score = -1.17, R) >consensus _______GUGA_AUAUGGC_CGCAUCCGUCGGAUGCGUGUUUCUAGAUGUCU_CUCUGUAUGUAUAUGCAUAGCCUGCCAAUGGCUAUG .................((.((((((.....)))))).))............................(((((((.......))))))) (-10.90 = -11.69 + 0.78)


| Location | 16,624,153 – 16,624,233 |
|---|---|
| Length | 80 |
| Sequences | 7 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 65.47 |
| Shannon entropy | 0.65469 |
| G+C content | 0.45962 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -7.66 |
| Energy contribution | -7.79 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16624153 80 - 27905053 CAUAGCCAUUGGCAGGCUAUGCAUAUACAUACAGAGAAGACAUCUAGUAACACGCAUCCGACGGAUGCG-GCCAUAU-UCAC------- (((((((.......)))))))........((((((.......))).)))...(((((((...)))))))-.......-....------- ( -22.20, z-score = -2.87, R) >droSim1.chr3R 22679596 79 + 27517382 CAUAGCCAUUGGCAGGCUAUGCAUAUACAUACAGAG-AGACAUCUAGAAACACGCAUCCGACGGAUGCG-GCCAUAU-UCAC------- (((((((.......)))))))...........(((.-.....))).......(((((((...)))))))-.......-....------- ( -20.80, z-score = -2.75, R) >droSec1.super_27 687535 79 + 799419 CAUAGCCAUUGGCAGGCUAUGCAUAUACAUACAGAG-AGACAUCUAGAAACACGCAUCCGACGGAUGCG-GCCAUAU-UCAC------- (((((((.......)))))))...........(((.-.....))).......(((((((...)))))))-.......-....------- ( -20.80, z-score = -2.75, R) >droYak2.chr3R 5419346 78 - 28832112 CAUAGCCCUCGGGGGGCUAUGCAUAUACAUACAGAG-AGACAUCUAG-AACACGCACCCGACGGAUGCG-GCCAUAU-UCAC------- ((((((((.....))))))))...........(((.-.....)))..-....((((((....)).))))-.......-....------- ( -20.60, z-score = -1.57, R) >droEre2.scaffold_4770 12706761 78 + 17746568 CAUAGCCCUUGGGCGGCUAUGCAUAUACAUACAGAG-AGACAUCUAG-AACACGCACCCGUUGGAUGCG-GACAUAU-UCAC------- ((((((((....).)))))))...........(((.-.....)))..-....((((((....)).))))-.......-....------- ( -16.70, z-score = -0.51, R) >droWil1.scaffold_181108 2466549 88 + 4707319 UACAUACAUAUG-UGUUUAUGCUUUGGCCUAUAGGCCAAGUUAAUAAACACAUAUGCUGAUUGAUUGUAUGUCGUGUGUCAUAUGUCAC .(((((((((((-(((((((..(((((((....)))))))...))))))))))))).((((((((.....))))...)))))))))... ( -30.90, z-score = -3.82, R) >droMoj3.scaffold_6540 29695606 77 - 34148556 CAUGUGCGGGGGCUCGUUAAGUAUGUAUACACUCAG-AUAU-UCUUGAUAUACUUGAUGAGAAUAAUCG-AUUG-GU-UCAC------- ((....(((...((((((((((((((..........-....-.....)))))))))))))).....)))-..))-..-....------- ( -16.61, z-score = -0.64, R) >consensus CAUAGCCAUUGGCAGGCUAUGCAUAUACAUACAGAG_AGACAUCUAGAAACACGCACCCGACGGAUGCG_GCCAUAU_UCAC_______ (((((((.......)))))))...............................((((.(.....).)))).................... ( -7.66 = -7.79 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:51 2011