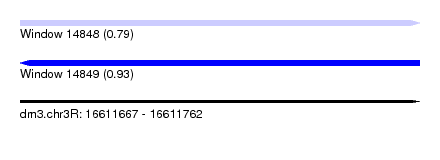
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,611,667 – 16,611,762 |
| Length | 95 |
| Max. P | 0.933342 |

| Location | 16,611,667 – 16,611,762 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 59.59 |
| Shannon entropy | 0.77537 |
| G+C content | 0.52697 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -5.31 |
| Energy contribution | -9.21 |
| Covariance contribution | 3.90 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
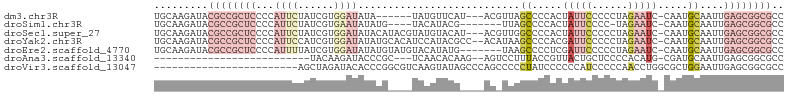
>dm3.chr3R 16611667 95 + 27905053 UGCAAGAUACGCCGCUCCCCAUUCUAUCGUGGAUAUA------UAUGUUCAU---ACGUUAGCCCCACUAUUCCCCCUAGAAUC-CAAUGCAAUUGAGCGGCGCC .........((((((((...........((((.....------.((((....---)))).....)))).((((......)))).-..........)))))))).. ( -22.50, z-score = -2.04, R) >droSim1.chr3R 22666437 92 - 27517382 UGCAAGAUACGCCGCUCCCCAUUCUAUCGUGAAUAUAUG----UACAUACG-------UUAGCCCCACUAUUCCCC-UAGAAUC-CAAUGCAAUUGAGCGGCGCC .........((((((((...((((......)))).....----........-------...((......((((...-..)))).-....))....)))))))).. ( -19.30, z-score = -1.76, R) >droSec1.super_27 675098 101 - 799419 UGCAAGAUACGCCGCUCCCCAUUCUAUCGUGGAUAUACAUACGUAUGUACAU---ACGUUGGCCCCACUAUUCCCCCUAGAAUC-CAAUGCAAUUGAGCGGCGCC .........((((((((.((((......))))((((((....))))))....---.((((((.......((((......)))))-))))).....)))))))).. ( -27.41, z-score = -2.35, R) >droYak2.chr3R 5404402 102 + 28832112 UGCAAGAUACGCCGCUCCCCAUUCCAUCGUGGAUAUAUGCACAUCCAUACGCC--ACAUAAGCCCCACGAUUCCCCCUAGAAUC-CAAUGCAAUUGAGCGGCGCC .........((((((((...........((((...((((......))))..))--))....((.....(((((......)))))-....))....)))))))).. ( -28.80, z-score = -3.75, R) >droEre2.scaffold_4770 12694555 97 - 17746568 UGCAAGAUACGCCGCUCCCCAUUUUAUCGUGGAUAUAUGUAUGUACAUAUG-------UAAGCCCCUCGAUUCCCCCUAGAAUC-CAAUGCAAUUGAGCGGCGCC .........((((((((.((((......))))((((((((....)))))))-------)..((.....(((((......)))))-....))....)))))))).. ( -29.50, z-score = -3.90, R) >droAna3.scaffold_13340 10726419 73 - 23697760 --------------------------UACAAGAUACCCGC---UCAACACAAG--AGUCCUUUACCGUUACUGCUCCCCACAUG-CGAUGCAAUUGAGCGGCGCC --------------------------..........((((---((((.....(--(((..............))))......((-(...))).)))))))).... ( -14.54, z-score = -1.06, R) >droVir3.scaffold_13047 7125703 81 - 19223366 ------------------------AGCUAGAUACACCCGGCGUCAAGUAUAGCCCAGCCCCCUAUCCCCCCAUCCCCCAACCUGGCGCUGGAAUUGAGCGGCGCC ------------------------..............((((((...((....(((((.((......................)).)))))...))...)))))) ( -17.25, z-score = 0.82, R) >consensus UGCAAGAUACGCCGCUCCCCAUUCUAUCGUGGAUAUACG____UACAUACA____AC_UUAGCCCCACUAUUCCCCCUAGAAUC_CAAUGCAAUUGAGCGGCGCC .........((((((((...((((......))))...........................((.....(((((......))))).....))....)))))))).. ( -5.31 = -9.21 + 3.90)
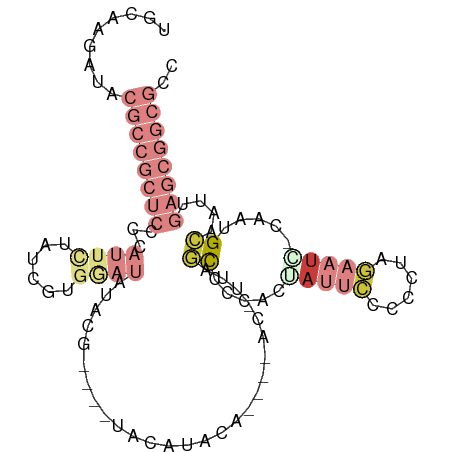
| Location | 16,611,667 – 16,611,762 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 59.59 |
| Shannon entropy | 0.77537 |
| G+C content | 0.52697 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -10.34 |
| Energy contribution | -13.41 |
| Covariance contribution | 3.07 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16611667 95 - 27905053 GGCGCCGCUCAAUUGCAUUG-GAUUCUAGGGGGAAUAGUGGGGCUAACGU---AUGAACAUA------UAUAUCCACGAUAGAAUGGGGAGCGGCGUAUCUUGCA .(((((((((.....((((.-.(((((....))))).(((((......((---((....)))------)...))))).....))))..)))))))))........ ( -28.30, z-score = -1.16, R) >droSim1.chr3R 22666437 92 + 27517382 GGCGCCGCUCAAUUGCAUUG-GAUUCUA-GGGGAAUAGUGGGGCUAA-------CGUAUGUA----CAUAUAUUCACGAUAGAAUGGGGAGCGGCGUAUCUUGCA .(((((((((.....(....-)((((((-..(((((((((..((...-------.)).....----))).))))).)..))))))...)))))))))........ ( -28.20, z-score = -1.63, R) >droSec1.super_27 675098 101 + 799419 GGCGCCGCUCAAUUGCAUUG-GAUUCUAGGGGGAAUAGUGGGGCCAACGU---AUGUACAUACGUAUGUAUAUCCACGAUAGAAUGGGGAGCGGCGUAUCUUGCA .(((((((((.....((((.-.(((((....))))).(((((....((((---((((....))))))))...))))).....))))..)))))))))........ ( -36.30, z-score = -2.30, R) >droYak2.chr3R 5404402 102 - 28832112 GGCGCCGCUCAAUUGCAUUG-GAUUCUAGGGGGAAUCGUGGGGCUUAUGU--GGCGUAUGGAUGUGCAUAUAUCCACGAUGGAAUGGGGAGCGGCGUAUCUUGCA .(((((((((.(((.((((.-((((((....))))))(((((...(((((--(.(((....)))))))))..))))))))).)))...)))))))))........ ( -42.70, z-score = -3.45, R) >droEre2.scaffold_4770 12694555 97 + 17746568 GGCGCCGCUCAAUUGCAUUG-GAUUCUAGGGGGAAUCGAGGGGCUUA-------CAUAUGUACAUACAUAUAUCCACGAUAAAAUGGGGAGCGGCGUAUCUUGCA .(((((((((....((.((.-((((((....)))))).))..))...-------.((((((....)))))).((((........)))))))))))))........ ( -33.30, z-score = -3.12, R) >droAna3.scaffold_13340 10726419 73 + 23697760 GGCGCCGCUCAAUUGCAUCG-CAUGUGGGGAGCAGUAACGGUAAAGGACU--CUUGUGUUGA---GCGGGUAUCUUGUA-------------------------- .((.((((((.(((((..(.-(....).)..)))))..........(((.--.....)))))---))))))........-------------------------- ( -20.40, z-score = 0.17, R) >droVir3.scaffold_13047 7125703 81 + 19223366 GGCGCCGCUCAAUUCCAGCGCCAGGUUGGGGGAUGGGGGGAUAGGGGGCUGGGCUAUACUUGACGCCGGGUGUAUCUAGCU------------------------ ....((.((((.(((((((.....)))))))..)))).))......(((((((.((((((((....)))))))))))))))------------------------ ( -31.50, z-score = -0.31, R) >consensus GGCGCCGCUCAAUUGCAUUG_GAUUCUAGGGGGAAUAGUGGGGCUAA_GU____UGUAUGUA____CAUAUAUCCACGAUAGAAUGGGGAGCGGCGUAUCUUGCA .(((((((((............(((((....))))).(((((..............................)))))...........)))))))))........ (-10.34 = -13.41 + 3.07)

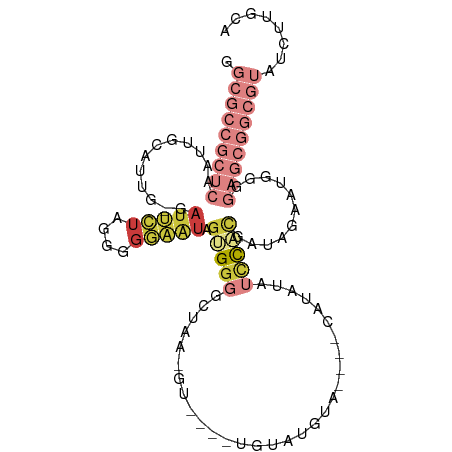
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:48 2011