
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,600,115 – 16,600,211 |
| Length | 96 |
| Max. P | 0.582914 |

| Location | 16,600,115 – 16,600,211 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 73.12 |
| Shannon entropy | 0.52144 |
| G+C content | 0.48171 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -11.35 |
| Energy contribution | -10.78 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
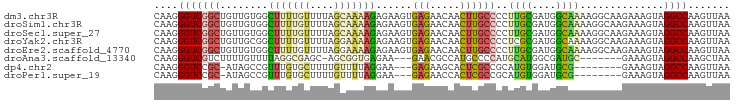
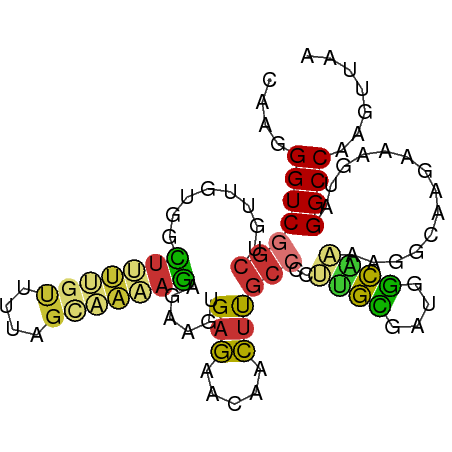
>dm3.chr3R 16600115 96 + 27905053 CAAGGGUCGGCUGUUGUGGCUUUUGUUUUAGCAAAAGAGAAGUGAGAACAACUUGCCCCUUGCGAUGGCAAAAGGCAAGAAAGUAGGCCAAGUUAA ....((((.((((((.(.((((((.(((.....))).)))))).).)))..((((((..((((....))))..))))))..))).))))....... ( -32.30, z-score = -2.34, R) >droSim1.chr3R 22654588 96 - 27517382 CAAGGGUCGGCUGUUGUGGCUUUUGUUUUAGCAAAAGAGAAGUGAGAACAACUUGCCCCUUGCGAUGGCAAAAGGCAAGAAAGUAGGCCAAGUUAA ....((((.((((((.(.((((((.(((.....))).)))))).).)))..((((((..((((....))))..))))))..))).))))....... ( -32.30, z-score = -2.34, R) >droSec1.super_27 663543 96 - 799419 CAAGGGUCGGCUGUUGUGGCUUUUGUUUUAGCAAAAGAGAAGUGAGAACAACUUGCCCCUUGCGAUGGCAAAAGGCAAGAAAGUAGGCCAAGUUAA ....((((.((((((.(.((((((.(((.....))).)))))).).)))..((((((..((((....))))..))))))..))).))))....... ( -32.30, z-score = -2.34, R) >droYak2.chr3R 5392534 96 + 28832112 CAAGGGUCGGCUGUUGCGGCUUUUGUUUUAGGAAAAGAGAAGUGAGAACAACUUGCCCCUCGCGAUGGCAAAAGGCAAGAAAGUAGGCCAAGUUAA ....((((.((((((.(.((((((.(((.....))).))))))..))))..((((((....((....))....))))))..))).))))....... ( -25.00, z-score = 0.12, R) >droEre2.scaffold_4770 12682975 96 - 17746568 CAAGGGUCGGCUGUUGUGGCUUUUGUUUUAGGAAAAGAGAAGUGAGAACAACUUGCCCCUUGCGAUGGCAAAAGGCAAGAAAGUAGGCCAAGUUAA ....((((.((((((.(.((((((.(((.....))).)))))).).)))..((((((..((((....))))..))))))..))).))))....... ( -32.30, z-score = -2.59, R) >droAna3.scaffold_13340 10714861 85 - 23697760 CAAGGGUCGUCUUUUGUUUUAGGCGAGC-AGCGGUGAGAA---GAACGCCAUGCCCCAUGCAUGGCGAUGC-------GAAAGUAGGCCAAGCUAA ...(((((((((........))))))((-(..((((....---...)))).))))))..((.((((..(((-------....))).)))).))... ( -32.70, z-score = -2.46, R) >dp4.chr2 5938316 84 + 30794189 CAAGGGUCCGC-AUAGCCGUUUGUGCUUUUGUUUUAGGAA---GAGAAGCACUCGCCGCAUGUGGAUGCG--------GAAAGUAGGCCAAGUUAA ...(.((((((-((.((.((..((((((((.(((...)))---.))))))))..)).)))))))))).)(--------(........))....... ( -29.70, z-score = -2.44, R) >droPer1.super_19 1674514 84 + 1869541 CAAGGGUCCGC-AUAGCCGUUUGUGCUUUUGUUUUAGGAA---GAGAACCACUCGCCGCAUGUGGAUGCG--------GAAAGUAGGCCAAGUUAA ...(.((((((-((.((.((..(((.((((.(((...)))---.)))).)))..)).)))))))))).)(--------(........))....... ( -23.10, z-score = -0.28, R) >consensus CAAGGGUCGGCUGUUGUGGCUUUUGUUUUAGCAAAAGAGAAGUGAGAACAACUUGCCCCUUGCGAUGGCAAAAGGCAAGAAAGUAGGCCAAGUUAA ....(((((((........(((((((....)))))))......(((.....))))))..((((....))))..............))))....... (-11.35 = -10.78 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:46 2011