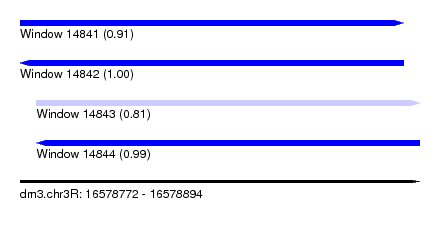
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,578,772 – 16,578,894 |
| Length | 122 |
| Max. P | 0.995665 |

| Location | 16,578,772 – 16,578,889 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.29 |
| Shannon entropy | 0.15208 |
| G+C content | 0.48466 |
| Mean single sequence MFE | -36.29 |
| Consensus MFE | -28.96 |
| Energy contribution | -30.00 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16578772 117 + 27905053 AUUCUUUUUGAAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCAGCCAAGGAUUUAUGCCCCUUUCGAGAAAGUCCUUUCAGU---GCUUGGCCGCUGUAUCGUUUACACUC .(((.....)))(((.(((.......))).)))(((.((.(((((((((((.((((((.(((.......((((....)))).......)))---.)))))).))))))))))).))))). ( -37.54, z-score = -2.87, R) >droEre2.scaffold_4770 12661970 117 - 17746568 AUUAUUUUUGGAGGCCUUAUGAAAUCGAUUGCUGGUUGUCAACGAUCCAGCGGCCAAGGACUCAUGCCCCUUUCGAGAACAUCCUCCCAAU---GCUUAGCCGCUGUAUCGUUUACACUC ..........((((((.................)))(((.((((((.(((((((.((((.........))))..(((......))).....---.....))))))).)))))).)))))) ( -29.23, z-score = -0.89, R) >droYak2.chr3R 5371136 120 + 28832112 AUUCUUUUUCGAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCGGCCAAGCACUCACGCCCCUUUCGAGAACGUUCUCACAAAACAGCUUAGCCGCUGUAUCGUUUACACAC ............(((.(((.......))).)))...(((.((((((((((((((.((((.(((...........)))...(((.......))).)))).)))))))))))))).)))... ( -38.90, z-score = -3.72, R) >droSec1.super_27 641979 117 - 799419 AUUCUUUUUGGAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCAGCCAAGGAUUCAUGCCCCUUUCGAGAACGUCCUCUCAGU---GCUUGGCCGCUGUAUCGUUUACACUC ..(((....)))(((.(((.......))).)))(((.((.(((((((((((.((((((.(((............((((......)))))))---.)))))).))))))))))).))))). ( -37.90, z-score = -2.19, R) >droSim1.chr3R 22633132 117 - 27517382 AUUCUUUUUGGAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCAGCCAAGGAUUCAUGCCCCUUUCGAGAACGUCCUCUCAGU---GCUUGGCCGCUGUAUCGUUUACACUC ..(((....)))(((.(((.......))).)))(((.((.(((((((((((.((((((.(((............((((......)))))))---.)))))).))))))))))).))))). ( -37.90, z-score = -2.19, R) >consensus AUUCUUUUUGGAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCAGCCAAGGAUUCAUGCCCCUUUCGAGAACGUCCUCUCAGU___GCUUGGCCGCUGUAUCGUUUACACUC ............(((.(((.......))).)))...(((.(((((((((((.((((((((............))(((......))).........)))))).))))))))))).)))... (-28.96 = -30.00 + 1.04)

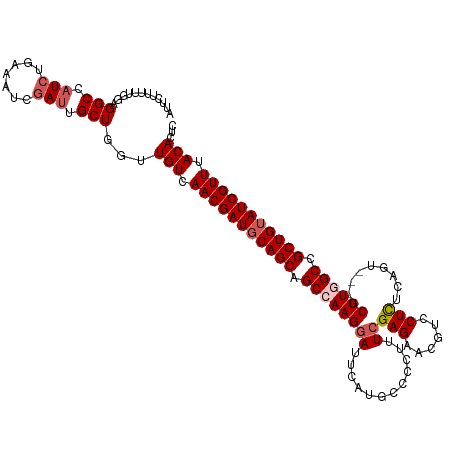
| Location | 16,578,772 – 16,578,889 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.29 |
| Shannon entropy | 0.15208 |
| G+C content | 0.48466 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -37.26 |
| Energy contribution | -36.78 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16578772 117 - 27905053 GAGUGUAAACGAUACAGCGGCCAAGC---ACUGAAAGGACUUUCUCGAAAGGGGCAUAAAUCCUUGGCUGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUUCAAAAAGAAU (..(((.((((((.(((((((((((.---.((....))....((((....))))........))))))))))).)))))).)))..).((.(((.......))).))............. ( -39.50, z-score = -3.47, R) >droEre2.scaffold_4770 12661970 117 + 17746568 GAGUGUAAACGAUACAGCGGCUAAGC---AUUGGGAGGAUGUUCUCGAAAGGGGCAUGAGUCCUUGGCCGCUGGAUCGUUGACAACCAGCAAUCGAUUUCAUAAGGCCUCCAAAAAUAAU ((((((.((((((.((((((((....---.(((((((....))))))).((((((....)))))))))))))).)))))).)))....((....(....).....))))).......... ( -37.30, z-score = -1.95, R) >droYak2.chr3R 5371136 120 - 28832112 GUGUGUAAACGAUACAGCGGCUAAGCUGUUUUGUGAGAACGUUCUCGAAAGGGGCGUGAGUGCUUGGCCGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUCGAAAAAGAAU ((.(((.((((((.((((((((((((.((((.....))))...(((....)))((....)))))))))))))).)))))).)))....))..((((..((....))..))))........ ( -46.30, z-score = -3.54, R) >droSec1.super_27 641979 117 + 799419 GAGUGUAAACGAUACAGCGGCCAAGC---ACUGAGAGGACGUUCUCGAAAGGGGCAUGAAUCCUUGGCUGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUCCAAAAAGAAU ((((((.((((((.(((((((((((.---.((....)).(((((((....)))).)))....))))))))))).)))))).)))....((.(((.......))).))))).......... ( -42.70, z-score = -3.45, R) >droSim1.chr3R 22633132 117 + 27517382 GAGUGUAAACGAUACAGCGGCCAAGC---ACUGAGAGGACGUUCUCGAAAGGGGCAUGAAUCCUUGGCUGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUCCAAAAAGAAU ((((((.((((((.(((((((((((.---.((....)).(((((((....)))).)))....))))))))))).)))))).)))....((.(((.......))).))))).......... ( -42.70, z-score = -3.45, R) >consensus GAGUGUAAACGAUACAGCGGCCAAGC___ACUGAGAGGACGUUCUCGAAAGGGGCAUGAAUCCUUGGCUGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUCCAAAAAGAAU (..(((.((((((.(((((((((((.....((....))....((((....))))........))))))))))).)))))).)))..).((.(((.......))).))............. (-37.26 = -36.78 + -0.48)

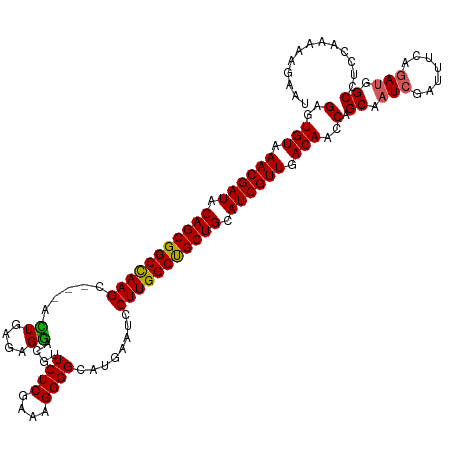
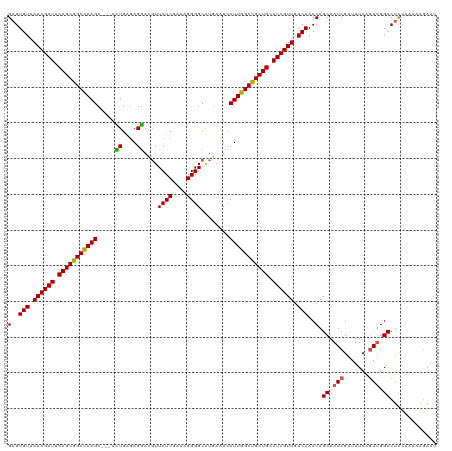
| Location | 16,578,777 – 16,578,894 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.03 |
| Shannon entropy | 0.15749 |
| G+C content | 0.49658 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -30.22 |
| Energy contribution | -31.26 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16578777 117 + 27905053 UUUUGAAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCAGCCAAGGAUUUAUGCCCCUUUCGAGAAAGUCCUUUCAGU---GCUUGGCCGCUGUAUCGUUUACACUCGUUGG .......(((.(((.......))).)))(..(((.(((((((((((.((((((.(((.......((((....)))).......)))---.)))))).))))))))))).)))..)..... ( -38.54, z-score = -2.47, R) >droEre2.scaffold_4770 12661975 117 - 17746568 UUUUGGAGGCCUUAUGAAAUCGAUUGCUGGUUGUCAACGAUCCAGCGGCCAAGGACUCAUGCCCCUUUCGAGAACAUCCUCCCAAU---GCUUAGCCGCUGUAUCGUUUACACUCGUUGA .....((((((.................)))(((.((((((.(((((((.((((.........))))..(((......))).....---.....))))))).)))))).))))))..... ( -30.93, z-score = -0.67, R) >droYak2.chr3R 5371141 120 + 28832112 UUUUCGAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCGGCCAAGCACUCACGCCCCUUUCGAGAACGUUCUCACAAAACAGCUUAGCCGCUGUAUCGUUUACACACGUUGU .......(((.(((.......))).))).(((((.((((((((((((((.((((.(((...........)))...(((.......))).)))).)))))))))))))).))).))..... ( -39.70, z-score = -3.02, R) >droSec1.super_27 641984 117 - 799419 UUUUGGAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCAGCCAAGGAUUCAUGCCCCUUUCGAGAACGUCCUCUCAGU---GCUUGGCCGCUGUAUCGUUUACACUCGUUGU .....(((((.(((.......))).))....(((.(((((((((((.((((((.(((............((((......)))))))---.)))))).))))))))))).))))))..... ( -39.60, z-score = -2.12, R) >droSim1.chr3R 22633137 117 - 27517382 UUUUGGAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCAGCCAAGGAUUCAUGCCCCUUUCGAGAACGUCCUCUCAGU---GCUUGGCCGCUGUAUCGUUUACACUCGUUGU .....(((((.(((.......))).))....(((.(((((((((((.((((((.(((............((((......)))))))---.)))))).))))))))))).))))))..... ( -39.60, z-score = -2.12, R) >consensus UUUUGGAGGCCAUCUGAAAUCGAUUGCUGGUUGUCAACGAUGCAGCAGCCAAGGAUUCAUGCCCCUUUCGAGAACGUCCUCUCAGU___GCUUGGCCGCUGUAUCGUUUACACUCGUUGU .......(((.(((.......))).)))(..(((.(((((((((((.((((((((............))(((......))).........)))))).))))))))))).)))..)..... (-30.22 = -31.26 + 1.04)

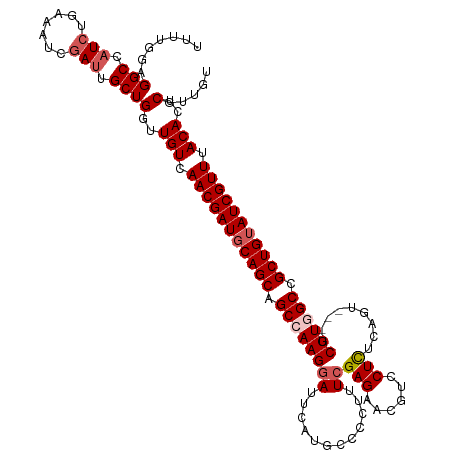
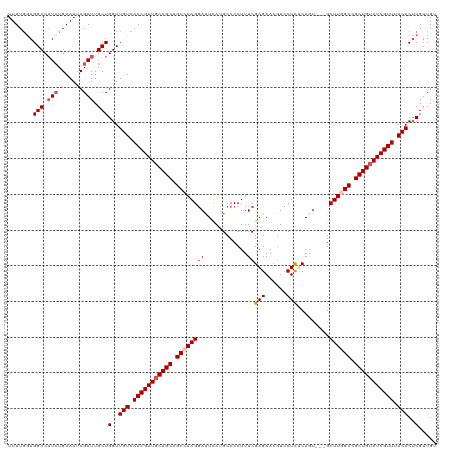
| Location | 16,578,777 – 16,578,894 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.03 |
| Shannon entropy | 0.15749 |
| G+C content | 0.49658 |
| Mean single sequence MFE | -42.06 |
| Consensus MFE | -37.56 |
| Energy contribution | -37.08 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16578777 117 - 27905053 CCAACGAGUGUAAACGAUACAGCGGCCAAGC---ACUGAAAGGACUUUCUCGAAAGGGGCAUAAAUCCUUGGCUGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUUCAAAA .....(..(((.((((((.(((((((((((.---.((....))....((((....))))........))))))))))).)))))).)))..).((.(((.......))).))........ ( -39.80, z-score = -3.34, R) >droEre2.scaffold_4770 12661975 117 + 17746568 UCAACGAGUGUAAACGAUACAGCGGCUAAGC---AUUGGGAGGAUGUUCUCGAAAGGGGCAUGAGUCCUUGGCCGCUGGAUCGUUGACAACCAGCAAUCGAUUUCAUAAGGCCUCCAAAA ....(((.(((.((((((.((((((((....---.(((((((....))))))).((((((....)))))))))))))).)))))).)))........))).................... ( -37.90, z-score = -1.68, R) >droYak2.chr3R 5371141 120 - 28832112 ACAACGUGUGUAAACGAUACAGCGGCUAAGCUGUUUUGUGAGAACGUUCUCGAAAGGGGCGUGAGUGCUUGGCCGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUCGAAAA .....((.(((.((((((.((((((((((((.((((.....))))...(((....)))((....)))))))))))))).)))))).)))....))..((((..((....))..))))... ( -46.60, z-score = -3.21, R) >droSec1.super_27 641984 117 + 799419 ACAACGAGUGUAAACGAUACAGCGGCCAAGC---ACUGAGAGGACGUUCUCGAAAGGGGCAUGAAUCCUUGGCUGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUCCAAAA .....((((((.((((((.(((((((((((.---.((....)).(((((((....)))).)))....))))))))))).)))))).)))....((.(((.......))).)))))..... ( -43.00, z-score = -3.39, R) >droSim1.chr3R 22633137 117 + 27517382 ACAACGAGUGUAAACGAUACAGCGGCCAAGC---ACUGAGAGGACGUUCUCGAAAGGGGCAUGAAUCCUUGGCUGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUCCAAAA .....((((((.((((((.(((((((((((.---.((....)).(((((((....)))).)))....))))))))))).)))))).)))....((.(((.......))).)))))..... ( -43.00, z-score = -3.39, R) >consensus ACAACGAGUGUAAACGAUACAGCGGCCAAGC___ACUGAGAGGACGUUCUCGAAAGGGGCAUGAAUCCUUGGCUGCUGCAUCGUUGACAACCAGCAAUCGAUUUCAGAUGGCCUCCAAAA .....(..(((.((((((.(((((((((((.....((....))....((((....))))........))))))))))).)))))).)))..).((.(((.......))).))........ (-37.56 = -37.08 + -0.48)

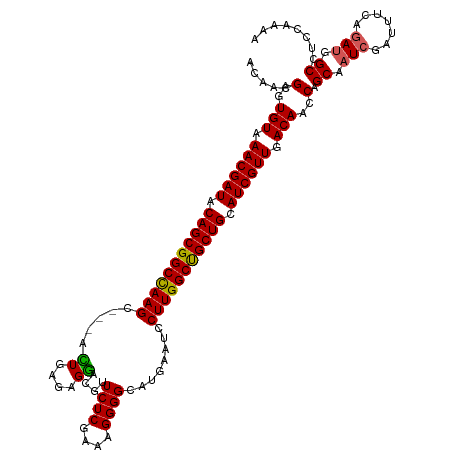
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:44 2011