
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,554,888 – 16,554,985 |
| Length | 97 |
| Max. P | 0.551823 |

| Location | 16,554,888 – 16,554,985 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.85 |
| Shannon entropy | 0.50700 |
| G+C content | 0.46862 |
| Mean single sequence MFE | -16.11 |
| Consensus MFE | -7.03 |
| Energy contribution | -6.73 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
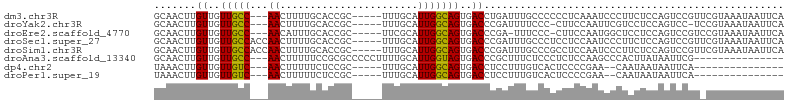
>dm3.chr3R 16554888 97 - 27905053 GCAACUUGUUGUUGCC---AACUUUUGCACCGC-----UUUGCAUUGGCAGUGACCUGAUUUGCCCCCCUCAAAUCCCUUCUCCAGUCCGUUCGUAAAUAAUUCA (((((..((..(((((---((....((((....-----..)))))))))))..))..((((((.......)))))).............))).)).......... ( -19.50, z-score = -2.25, R) >droYak2.chr3R 5341367 95 - 28832112 GCAACUUGUUGUUGCC---AACUUUUGCACCGC-----UUUGCAUUGGCAGUGACCCGAUUUUCCC-CUUCCAAUUCGUCCUCCAGUCC-UCCGUAAAUAAUUCA .......((..(((((---((....((((....-----..)))))))))))..))...........-......................-............... ( -15.70, z-score = -1.44, R) >droEre2.scaffold_4770 12636699 95 + 17746568 GCAACUUGUUGUUGCC---AACAUUUGCACCGC-----UUCGCAUUGGCAGUGACCCGA-UUUCCC-CUUCCAAUGGCUCCUCCAGUCCGUCCGUAAAUAAUUCA ((((..(((((....)---)))).))))..((.-----..((.(((((.((.((.(((.-......-.......))).))))))))).))..))........... ( -17.54, z-score = -0.61, R) >droSec1.super_27 617859 100 + 799419 GCAACUUGUUGUUGCCACCAACUUUUGCACCGC-----UUUGCAUUGGCAGUGACCCGAUUUGCCCUCCUCCAAUCCCUUCUCCAGUCCGUUCGUAAAUAAUUCA ((((.((((..((((((........((((....-----..)))).))))))..))..)).))))......................................... ( -16.00, z-score = -0.91, R) >droSim1.chr3R 22608285 100 + 27517382 GCAACUUGUUGUUGCCACCAACUUUUGCACCGC-----UUUGCAUUGGCAGUGACCCGAUUUGCCCGCCUCCAAUCCCUUCUCCAGUCCGUUCGUAAAUAAUUCA ((((.((((..((((((........((((....-----..)))).))))))..))..)).))))......................................... ( -16.00, z-score = -0.27, R) >droAna3.scaffold_13340 10666828 87 + 23697760 GCAACUUGUUGUUGCC---AACUUUUUCCGCGCCCCCUUUUGCAUUGGUAGUGACCCGCUUUCUCCCUCUCCAAGCCCACUUAUAAUUCG--------------- (((((.....))))).---...........(((..((.........))..)))....((((...........))))..............--------------- ( -10.90, z-score = -0.53, R) >dp4.chr2 5890269 80 - 30794189 UAAACUUGUUGUUGUC---AACUUUUUCUCCGC-----UUUGCAUUGGCAGUGACCUCCUUUGUCACUCCCCGAA--CAAUAAUAAUUCA--------------- .....(((((((((((---............((-----...))...((.((((((.......)))))).)).).)--)))))))))....--------------- ( -16.60, z-score = -3.23, R) >droPer1.super_19 1622261 80 - 1869541 UAAACUUGUUGUUGUC---AACUUUUUCUCCGC-----UUUGCAUUGGCAGUGACCUCCUUUGUCACUCCCCGAA--CAAUAAUAAUUCA--------------- .....(((((((((((---............((-----...))...((.((((((.......)))))).)).).)--)))))))))....--------------- ( -16.60, z-score = -3.23, R) >consensus GCAACUUGUUGUUGCC___AACUUUUGCACCGC_____UUUGCAUUGGCAGUGACCCGAUUUGCCCCCCUCCAAUCCCUCCUCCAGUCCGU_CGUAAAUAAUUCA .......((..(((((..............................)))))..)).................................................. ( -7.03 = -6.73 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:39 2011