| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,550,922 – 16,551,017 |
| Length | 95 |
| Max. P | 0.983169 |

| Location | 16,550,922 – 16,551,017 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Shannon entropy | 0.48291 |
| G+C content | 0.40808 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -12.81 |
| Energy contribution | -14.27 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
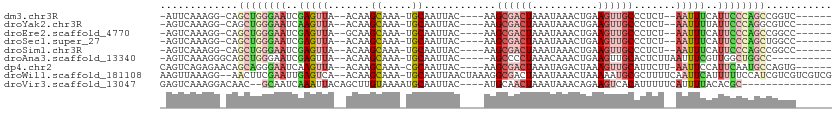
>dm3.chr3R 16550922 95 + 27905053 -AUUCAAAGG-CAGCUGGGAAUCGAGUUA--ACAAGCAAA-UGCAAUUAC----AAGCGACUAAAUAAACUGAAGUUGCCCUCU--AAUUUCAUUCCCAGCCGGUC------ -.......((-(.(((((((((.((((((--....((...-.))......----..((((((...........))))))....)--))))).)))))))))..)))------ ( -27.60, z-score = -3.38, R) >droYak2.chr3R 5337328 95 + 28832112 -AGUCAAAGG-CAGCUGGGAAUCAAGUUA--ACAAGCAAA-UGCAAUUAC----AAGCGACUAAAUAAACUGAAGUUGCCCUCU--AAUUUUAUUCCCAGGCGUCC------ -.......((-(..((((((((.((((((--....((...-.))......----..((((((...........))))))....)--))))).))))))))..))).------ ( -24.80, z-score = -2.45, R) >droEre2.scaffold_4770 12632713 95 - 17746568 -AGUCAAAGG-CAGCUGGGAAUCGAGUUA--GCAAGCAAA-UGCAAUUAC----AAGCGACUAAAUAAACUGAAGUUGCCCUCU--AAUUUCAUUCCCAGCCGGCC------ -.......((-(.(((((((((.((((((--(...((...-.))......----..((((((...........))))))...))--))))).)))))))))..)))------ ( -32.50, z-score = -4.05, R) >droSec1.super_27 613951 95 - 799419 -AGUCAAAGG-CAGCUGGGAAUCGAGUUA--ACAAGCAAA-UGCAAUUAC----AAGCGACUAAAUAAACUGAAGUUGCCCUCU--AAUUUCAUUCCCAGCUGGCC------ -.......(.-(((((((((((.((((((--....((...-.))......----..((((((...........))))))....)--))))).))))))))))).).------ ( -32.30, z-score = -4.29, R) >droSim1.chr3R 22604335 95 - 27517382 -AGUCAAAGG-CAGCUGGGAAUCGAGUUA--ACAAGCAAA-UGCAAUUAC----AAGCGACUAAAUAAACUGAAGUUGCCCUCU--AAUUUCAUUCCCAGCCGGCC------ -.......((-(.(((((((((.((((((--....((...-.))......----..((((((...........))))))....)--))))).)))))))))..)))------ ( -30.30, z-score = -3.83, R) >droAna3.scaffold_13340 10662532 93 - 23697760 -AGUCAAAGGGCAGCUGGGAAUCGAGUUA--ACAAGCAAA-UGCAAUUAC-----AGCCCCUAAACAAACUGAAGUUGCACUCUUAAUUUCGUUGGCUGGCC---------- -.......((.((((..((((....(((.--...)))...-(((((((.(-----((............))).)))))))........))).)..)))).))---------- ( -22.20, z-score = -0.02, R) >dp4.chr2 5886394 98 + 30794189 CAGUCAGAGAACAGCAGGGAAUCAAGUUA--ACAAGCAAA-CGCAAUUAC----AAGCGACUAAAUAGACUAAAGUUGCAUUCUU-AAUUCCAUUCAAUGCCAGUG------ .............(((..((((..(((((--(...((...-.))......----..((((((...........))))))....))-))))..))))..))).....------ ( -16.80, z-score = -0.99, R) >droWil1.scaffold_181108 2389456 107 - 4707319 AAGUUAAAGG--AACUUCGAAUUGAGUCA--ACAAGCAAA-UGCAAUUAACUAAAGGCGACUAAAUAAACUAAAAAUGCGCUUUUCAAUUCAUUUUUCCAUCGUCGUCGUCG ........((--((....(((((((....--....((...-.)).........((((((.(................))))))))))))))....))))............. ( -16.59, z-score = -0.37, R) >droVir3.scaffold_13047 7064423 91 - 19223366 GAGUCAAAGGACAAC--GCAAUCAAAUUACAGCUUGUAAAAUGCAAUUAC----AUGCAACUAAAUAAACAGAAGUCACAUUUUUCAUUUUACACGC--------------- ..(((....)))...--..............((.(((((((((((.....----.))).............((((.......)))))))))))).))--------------- ( -13.20, z-score = -1.46, R) >consensus _AGUCAAAGG_CAGCUGGGAAUCGAGUUA__ACAAGCAAA_UGCAAUUAC____AAGCGACUAAAUAAACUGAAGUUGCCCUCU__AAUUUCAUUCCCAGCCGGCC______ .............((((((((..(((((.......((.....))............((((((...........)))))).......)))))..))))))))........... (-12.81 = -14.27 + 1.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:37 2011