
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,524,937 – 16,525,036 |
| Length | 99 |
| Max. P | 0.929565 |

| Location | 16,524,937 – 16,525,036 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.49493 |
| G+C content | 0.53476 |
| Mean single sequence MFE | -28.61 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16524937 99 + 27905053 UAAUCGAUGCCAAC-AGCCUCGACUCGCUAACCGGGUUUCAGCCU----GGCAGCGGGCUGAAUGCCAGCCAUUCAAACGUGGCCCAUUCAACUGCACUCAGCC- ...((((.((....-.)).))))..((((..((((((....))))----)).))))((((((.(((..(((((......)))))..........))).))))))- ( -35.30, z-score = -2.36, R) >droWil1.scaffold_181108 2354427 100 - 4707319 UAAUCGAUGGCAGC--UCCACAGUAAAUCGAAUUUCAAUAAAGUCCAC-GAUGGC-AGCCGAAGGC-AACCAUUCAAACGUAACCCAUUCAAUUGCAGUUCACUC .....(((....((--(....)))..)))(((((.((((...(....)-(((((.-.(((...)))-..))))).................)))).))))).... ( -13.30, z-score = 0.74, R) >droAna3.scaffold_13340 10637321 105 - 23697760 UAAUCGAUAAUUCCGAGUUUCAAUACGUUGGCUGCCUUACGGGCUUACAGGAAACGAGCUGAAUGCCAGGCAUUCAAACGUGGCCCAUUCAGUUGCACUCAGCCG ...(((.......))).............(((((((....))((.....(....).((((((((((((.(........).))))..))))))))))...))))). ( -27.80, z-score = -0.56, R) >droEre2.scaffold_4770 12605742 99 - 17746568 UAAUCGAUGCCUAC-AGCCUCGACUCGCUAACCGGGUUUCAGCAU----GGUAGCGGGCUGAAUGCCAGCCAUUCAAACGUGGCCCAUUCAACUGCACUCAGCC- ...((((.((....-.)).))))..(((((.(((.((....)).)----)))))))((((((.(((..(((((......)))))..........))).))))))- ( -31.60, z-score = -1.42, R) >droYak2.chr3R 5313503 90 + 28832112 UAAUCGAUGCUUAC-AGCCUCGACUCGCUUUCC---------CAU----GGCAGCGGGCUGAAUGCCAGCCAUUCAAACGUGGCCCAUUCAACUGCACUCAGCC- ...((((.((....-.)).))))...(((....---------...----))).((((..((((((...(((((......))))).)))))).))))........- ( -26.50, z-score = -1.37, R) >droSec1.super_27 587630 99 - 799419 UAAUCGAUGCAAAC-AGCCUCGACUCGCUAACCGGGUUUCAGCCA----GGCAGCGGGCUGAAUGCCAGCCAUUCAAACGUGGCCCAUUUAACUGCACUCAGCC- ...((((.((....-.)).))))..((((..((.(((....))).----)).))))((((((.(((..(((((......)))))..........))).))))))- ( -31.90, z-score = -1.56, R) >droSim1.chr3R 22578399 99 - 27517382 UAAUCGAUGCCAAC-AGCCUCGACUCGCUAACCGGGUUUCAUCCU----GGCAGCGGGCUGAAUGCCAGCCAUUCAAACGUGGCCCAUUCAACUGCACUCAGCC- ...((((.((....-.)).))))..((((..(((((......)))----)).))))((((((.(((..(((((......)))))..........))).))))))- ( -33.90, z-score = -2.46, R) >consensus UAAUCGAUGCCAAC_AGCCUCGACUCGCUAACCGGGUUUCAGCCU____GGCAGCGGGCUGAAUGCCAGCCAUUCAAACGUGGCCCAUUCAACUGCACUCAGCC_ ...((((.((......)).))))..((((.......................))))((((((.(((..(((((......)))))..........))).)))))). (-14.86 = -15.40 + 0.54)

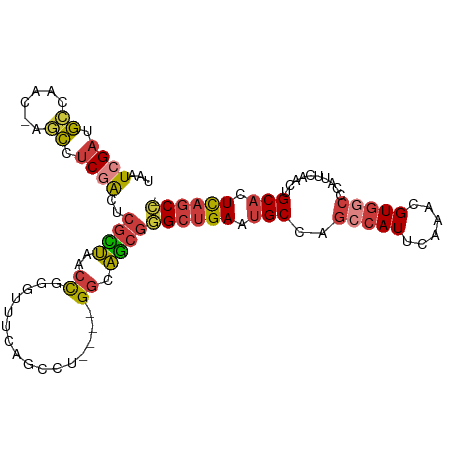
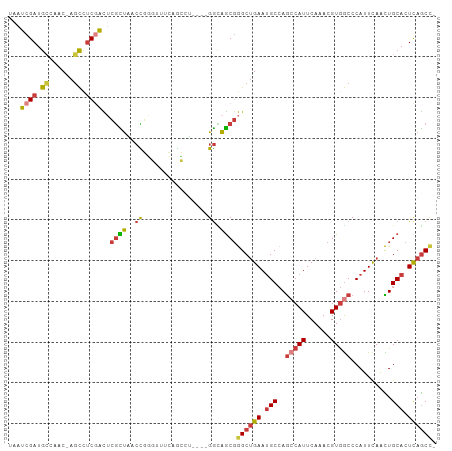
| Location | 16,524,937 – 16,525,036 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.49493 |
| G+C content | 0.53476 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -24.64 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16524937 99 - 27905053 -GGCUGAGUGCAGUUGAAUGGGCCACGUUUGAAUGGCUGGCAUUCAGCCCGCUGCC----AGGCUGAAACCCGGUUAGCGAGUCGAGGCU-GUUGGCAUCGAUUA -(((((((((((((((((((.....)))))....)))).))))))))))(((((((----.((......)).)).)))))((((((.((.-....)).)))))). ( -41.40, z-score = -1.83, R) >droWil1.scaffold_181108 2354427 100 + 4707319 GAGUGAACUGCAAUUGAAUGGGUUACGUUUGAAUGGUU-GCCUUCGGCU-GCCAUC-GUGGACUUUAUUGAAAUUCGAUUUACUGUGGA--GCUGCCAUCGAUUA .(((((((..(..(..((((.....))))..)(((((.-(((...))).-))))).-)..)......(((.....)))))))))((((.--....))))...... ( -21.90, z-score = 0.47, R) >droAna3.scaffold_13340 10637321 105 + 23697760 CGGCUGAGUGCAACUGAAUGGGCCACGUUUGAAUGCCUGGCAUUCAGCUCGUUUCCUGUAAGCCCGUAAGGCAGCCAACGUAUUGAAACUCGGAAUUAUCGAUUA .(((((..((((...((((((((......(((((((...)))))))))))))))..))))...((....))))))).............((((.....))))... ( -28.70, z-score = -0.01, R) >droEre2.scaffold_4770 12605742 99 + 17746568 -GGCUGAGUGCAGUUGAAUGGGCCACGUUUGAAUGGCUGGCAUUCAGCCCGCUACC----AUGCUGAAACCCGGUUAGCGAGUCGAGGCU-GUAGGCAUCGAUUA -(((((((((((((((((((.....)))))....)))).))))))))))(((((..----..((((.....)))))))))((((((.((.-....)).)))))). ( -38.40, z-score = -1.63, R) >droYak2.chr3R 5313503 90 - 28832112 -GGCUGAGUGCAGUUGAAUGGGCCACGUUUGAAUGGCUGGCAUUCAGCCCGCUGCC----AUG---------GGAAAGCGAGUCGAGGCU-GUAAGCAUCGAUUA -(((((((((((((((((((.....)))))....)))).))))))))))((((.((----...---------))..))))((((((.((.-....)).)))))). ( -34.80, z-score = -1.45, R) >droSec1.super_27 587630 99 + 799419 -GGCUGAGUGCAGUUAAAUGGGCCACGUUUGAAUGGCUGGCAUUCAGCCCGCUGCC----UGGCUGAAACCCGGUUAGCGAGUCGAGGCU-GUUUGCAUCGAUUA -(((((((((((((((((((.....))))))....))).))))))))))(((((((----.((......)).)).)))))((((((.((.-....)).)))))). ( -41.70, z-score = -2.37, R) >droSim1.chr3R 22578399 99 + 27517382 -GGCUGAGUGCAGUUGAAUGGGCCACGUUUGAAUGGCUGGCAUUCAGCCCGCUGCC----AGGAUGAAACCCGGUUAGCGAGUCGAGGCU-GUUGGCAUCGAUUA -(((((((((((((((((((.....)))))....)))).))))))))))(((((((----.((......)).)).)))))((((((.((.-....)).)))))). ( -41.40, z-score = -2.30, R) >consensus _GGCUGAGUGCAGUUGAAUGGGCCACGUUUGAAUGGCUGGCAUUCAGCCCGCUGCC____AGGCUGAAACCCGGUUAGCGAGUCGAGGCU_GUUGGCAUCGAUUA .(((((((((((((((((((.....)))))....)))).))))))))))((((((..................)).))))((((((.((......)).)))))). (-24.64 = -24.96 + 0.32)

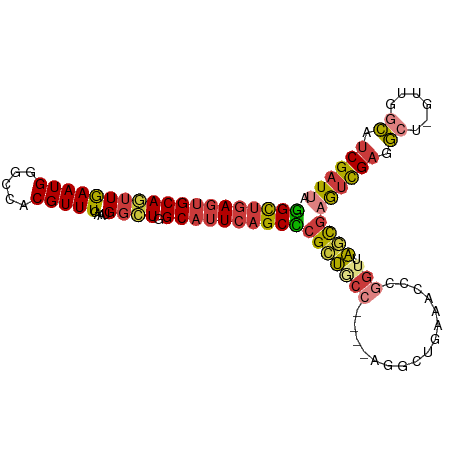
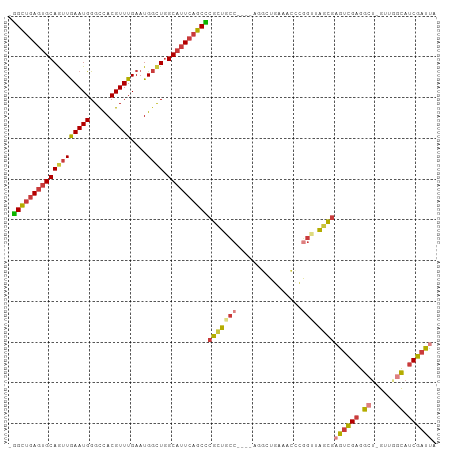
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:33 2011