
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,501,654 – 16,501,752 |
| Length | 98 |
| Max. P | 0.840795 |

| Location | 16,501,654 – 16,501,752 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.44758 |
| G+C content | 0.36836 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840795 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 16501654 98 + 27905053 ------CUAGGGAAAAGCAUAUGUAAAGUACAUUGCCUACUUUUGGGCAUAA-------UUUAUGCAUUACUC--AACCCGACAUUCAAAAGCUACUGUC---AGAGCUUUUGUAC ------...(((...((...(((((((((....((((((....))))))..)-------))).)))))..)).--..)))......((((((((......---..))))))))... ( -23.20, z-score = -1.77, R) >droSim1.chr3R 22555093 95 - 27517382 ------CUAGGGAAA-GCAUAUGUAAU-UACAUUGC-UACUUUGGGGCAUAA-------UUUAUGCAUUACUC--AACCCGACAUUCAAAAGCUGCUGUC---AGAGCUUUUGUAC ------...(((..(-(((.((((...-.)))))))-)...((((((((((.-------..)))))....)))--)))))......((((((((......---..))))))))... ( -24.10, z-score = -1.90, R) >droSec1.super_27 564920 98 - 799419 ------CUAGGGAAAAGCAUAUGUAAAUUACAUUGCCUACUUUUAGGCAUAA-------UUUAUGCAUUACUC--AACCCGACAUUCAAAAGCUGCUGUC---AGAGCUUUUGUAC ------...(((...((...((((((((((...((((((....)))))))))-------))..)))))..)).--..)))......((((((((......---..))))))))... ( -23.50, z-score = -2.14, R) >droYak2.chr3R_random 2812028 98 + 3277457 ------UUAGGGAAAAGCAUAUGUAAAUUACAUUGCCUACUUUUAGGCAUAA-------UUUAUGCAUUACUU--AAACCGACAUUCGAAAGCUGCUGUG---UGUGCUUUUGUAC ------....(.((((((((((((((((((...((((((....)))))))))-------)))))(((...(((--....((.....)).))).)))...)---))))))))).).. ( -22.90, z-score = -1.47, R) >droEre2.scaffold_4770 12582315 91 - 17746568 ------CUAGGGAAAAGCAUAUGUAAAUUACAUUGCCUACUUUUGGGCAUAA-------UUUAUGCAUUACUU--AACCCGACAUUCGAAAGC----------UGAGCUUUUGUAC ------...(((..(((...((((((((((...((((((....)))))))))-------))..)))))..)))--..)))......(((((((----------...)))))))... ( -21.50, z-score = -2.36, R) >droAna3.scaffold_13340 10614739 100 - 23697760 ------CUAGUAAAGAGCAUAUGUAAAUGGCAGUGCCUACUUUCGGGCAUAA-------UUUAUGCACCGAUUCCGACCUGACAUUUGAAAGCUGCUGUC---AAGCUUUUUAGAA ------....((((((((..((....))((((((((......(((((((((.-------..))))).))))...(((........)))...)).))))))---..))))))))... ( -23.70, z-score = -0.73, R) >droWil1.scaffold_181108 2329975 114 - 4707319 UGAUGAGAUGCAAAUAGAUAAUGUAAUUUCCAUAACCUACUUUUAGGCAUAAAAUAUAAUUUAUGCAAAAAUA--AAGUUGCCAGCUGGCAGCUGCUGUCCAAAAAGCUCUUGUAA ..(..(((.((.....((((..........................(((((((......))))))).......--.(((((((....)))))))..))))......)))))..).. ( -23.80, z-score = -0.83, R) >consensus ______CUAGGGAAAAGCAUAUGUAAAUUACAUUGCCUACUUUUGGGCAUAA_______UUUAUGCAUUACUC__AACCCGACAUUCGAAAGCUGCUGUC___AGAGCUUUUGUAC ................(((((............((((((....))))))............)))))....................((((((((...........))))))))... (-13.77 = -14.35 + 0.58)

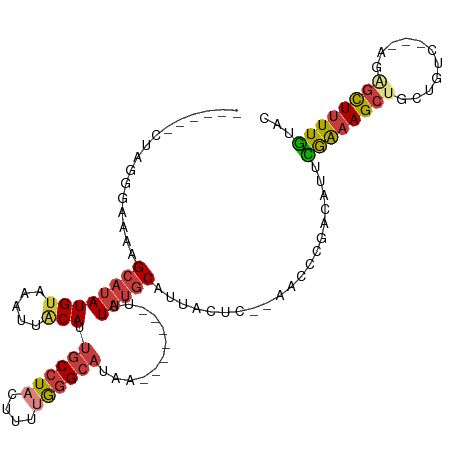
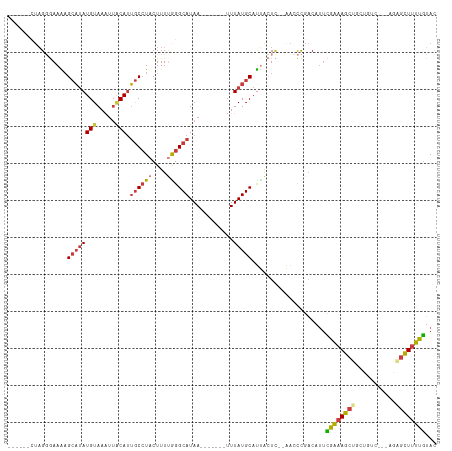
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:32 2011