| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,496,146 – 16,496,237 |
| Length | 91 |
| Max. P | 0.991297 |

| Location | 16,496,146 – 16,496,237 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 58.64 |
| Shannon entropy | 0.82620 |
| G+C content | 0.49944 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -6.88 |
| Energy contribution | -6.78 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
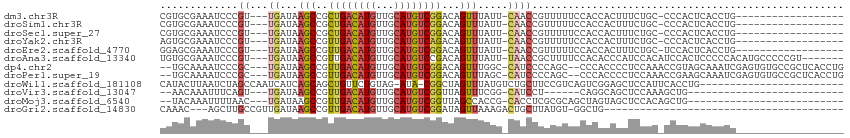
>dm3.chr3R 16496146 91 + 27905053 CGUGCGAAAUCCCGU---UGAUAAGCCGCUGACAUGUUGCAUGUCGGACAGUUUAUU-CAACCGUUUUUCCACCACUUUCUGC-CCCACUCACCUG------------------ .(((.((((..(.((---((((((((.(((((((((...)))))))).).))))).)-)))).)..)))))))..........-............------------------ ( -26.00, z-score = -4.05, R) >droSim1.chr3R 22549634 91 - 27517382 CGUGCGAAAUCCCGU---UGAUAAGCCGCUGACAUGUUGCAUGUCGGACAGUUUAUU-CAACCGUUUUUCCACCACUUUCUGC-CCCACUCACCUG------------------ .(((.((((..(.((---((((((((.(((((((((...)))))))).).))))).)-)))).)..)))))))..........-............------------------ ( -26.00, z-score = -4.05, R) >droSec1.super_27 559840 91 - 799419 CGUGCGAAAUCCCGU---UGAUAAGCCGCUGACAUGUUGCAUGUCGGACAGUUUAUU-CAACCGUUUUUCCACCACUUUCUGC-CCCACUCACCUG------------------ .(((.((((..(.((---((((((((.(((((((((...)))))))).).))))).)-)))).)..)))))))..........-............------------------ ( -26.00, z-score = -4.05, R) >droYak2.chr3R 5291174 91 + 28832112 AGUGCGAAAUCCCGU---UGAUAAGCCGUUGACAUGUUGCAUGUCAGACAGUUUAUU-CAACCGUUUUUCCACCACUUUCUGC-CCCACUCACCUG------------------ .(((.((((..(.((---((((((((.(((((((((...))))))).)).))))).)-)))).)..)))))))..........-............------------------ ( -24.20, z-score = -4.30, R) >droEre2.scaffold_4770 12576639 91 - 17746568 GGAGCGAAAUCCCGU---UGAUAAGCCGUUGACAUGUUGCAUGUCGGACAGUUUAUU-CAACCGUUUUUCCACCACUUUCUGC-UCCACUCACCUG------------------ (((((((((..(.((---((((((((.(((((((((...)))))).))).))))).)-)))).)..))))...........))-))).........------------------ ( -27.00, z-score = -3.78, R) >droAna3.scaffold_13340 10609247 103 - 23697760 UGUGCGAAAUCCCGU---UGAUAAGCCGUUGACAUGUUGCAUGUCCGACAGUUUAUU-UAACCGCUUUUCCACACCCAUCCACAUCCACUCCCCCACAUGCCCCCGU------- ((((.((((..(.((---((((((((.(((((((((...))))).)))).))))).)-)))).)..)))))))).................................------- ( -21.80, z-score = -3.28, R) >dp4.chr2 5830448 106 + 30794189 --UGCAAAAUCCCGC---UGAUAAGCCGUUGACAUGUUGCAUGUCGGACAGUUUGGC-CAUCCCCAGC--CCCACCCCUCCAAACCGUAGCAAAUCGAGUGUGCCGCUCACCUG --(((......((((---(....)))....((((((...))))))))((.((((((.-..........--.........)))))).)).)))....(((((...)))))..... ( -23.25, z-score = -0.51, R) >droPer1.super_19 1560963 106 + 1869541 --UGCAAAAUCCCGC---UGAUAAGCCGUUGACAUGUUGCAUGUCGGACAGUUUAGC-CAUCCCCAGC--CCCACCCCUCCAAACCGAAGCAAAUCGAGUGUGCCGCUCACCUG --(((........((---((.(((((.(((((((((...)))))).))).)))))..-......))))--........((......)).)))....(((((...)))))..... ( -20.00, z-score = -0.37, R) >droWil1.scaffold_181108 2324337 88 - 4707319 CAUACUUAAUCUAGCCAAUCAUCAGCAGCUGUUCUGUAG-AUA-CGGCUAGUUUAUGUCUGCUUCCGUCAGUCGGAGCUCCAUUCACCUG------------------------ ...((.(((.((((((....(((.((((.....)))).)-)).-.)))))).))).))..(((.(((.....))))))............------------------------ ( -21.50, z-score = -1.83, R) >droVir3.scaffold_13047 6989658 76 - 19223366 --AACAAAUUUCAGU---UGAUAAGCCGUUGACAUGUUGCAUGUCGGUUAGUUUCGG-CAUCCU------CAGGCAGCUCCAAAGCUG-------------------------- --...........((---(((..((((...((((((...))))))))))....))))-).....------....(((((....)))))-------------------------- ( -16.90, z-score = -0.25, R) >droMoj3.scaffold_6540 29570071 82 + 34148556 --UACAAAUUUUAAC---UGAUAAGCCGUUGACAUGUUGCAUGUCGGUUAGCCACCG-CACCUCGCGCAGCUAGUAGCUCCACAGCUG-------------------------- --........(((((---(((((.((..(......)..)).))))))))))....((-(.....)))(((((.((......)))))))-------------------------- ( -18.20, z-score = 0.17, R) >droGri2.scaffold_14830 937603 70 + 6267026 CAAAC---AGCUUGCCGUUGAUAAGCCGUUGACAUGUUGCAUGUCGGAUAGUUAAAGACUGCUUAUGU-GGCUG---------------------------------------- ....(---((((.((.(((....(((..((((((((...))))))))...)))...))).))......-)))))---------------------------------------- ( -17.00, z-score = -0.33, R) >consensus CGUGCGAAAUCCCGU___UGAUAAGCCGUUGACAUGUUGCAUGUCGGACAGUUUAUU_CAACCGUUUUUCCACCACUCUCCAC_CCCACUCACCUG__________________ .......................(((..((((((((...))))))))...)))............................................................. ( -6.88 = -6.78 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:29 2011