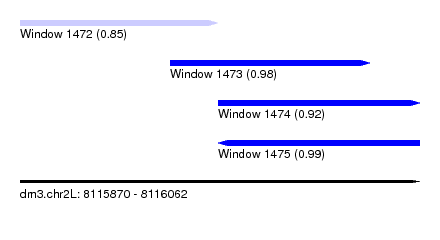
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,115,870 – 8,116,062 |
| Length | 192 |
| Max. P | 0.987490 |

| Location | 8,115,870 – 8,115,965 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Shannon entropy | 0.44139 |
| G+C content | 0.30211 |
| Mean single sequence MFE | -16.15 |
| Consensus MFE | -8.21 |
| Energy contribution | -9.09 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848690 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
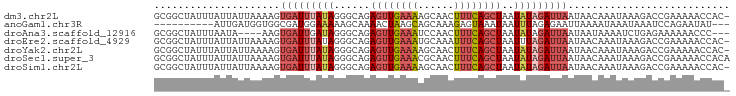
>dm3.chr2L 8115870 95 + 23011544 GCGGCUAUUUAUUAUUAAAAGUGAUUUAUAGGGCAGAGUUGAAAAGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCAC- .(((...((((((........(((((((((.((((((((((.....)))))))..)))..)))))))))......))))))..))).........- ( -18.94, z-score = -2.94, R) >anoGam1.chr3R 42862608 83 - 53272125 ----------AUUGAUGGUGGCGAUGGAAAAAGCAAAACUAAGCAGCAAAGAGUAAAUAAUUUAGAGAAUUAAAAUAAAUAAAUCCAGAAUAU--- ----------..............((((....((........)).............((((((...))))))...........))))......--- ( -5.50, z-score = 0.85, R) >droAna3.scaffold_12916 11839311 89 - 16180835 GCGGCUAUUUAAUA----AAGUGAUUGAUAGGGCAGAGUUGAAAUCCAACUUUCAGCUAAUAUAGAUUAAUAAUAAAAUCUGAGAAAAAACCC--- ...(((..(((((.----.....)))))...)))..((((((((......))))))))....((((((........))))))...........--- ( -14.80, z-score = -1.07, R) >droEre2.scaffold_4929 17021607 95 + 26641161 GCGGCUAUUUAUUAUUAAAAGUGAUUUAUAGGGCAGAGUUGAAAUGCAAAUUUCAGCUAAUUUAGAUUAAUAACAAAUAAAGACCGAAAAACCAC- .(((((((..(((((.....)))))..)))).....(((((((((....))))))))).........................))).........- ( -17.00, z-score = -1.88, R) >droYak2.chr2L 10746730 95 + 22324452 GCGGCUAUUUAUUAUUAAAAGUGAUUUAUAGGGCAGAGUUGAAAAGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCAC- .(((...((((((........(((((((((.((((((((((.....)))))))..)))..)))))))))......))))))..))).........- ( -18.94, z-score = -2.94, R) >droSec1.super_3 3612245 96 + 7220098 GCGGCUAUUUAUUAUUAAAAGUGAUUUAUAGGGCAGAGUUGAAACGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCACA .(((...((((((........(((((((((.((((((((((.....)))))))..)))..)))))))))......))))))..))).......... ( -18.94, z-score = -2.80, R) >droSim1.chr2L 7895383 95 + 22036055 GCGGCUAUUUAUUAUUAAAAGUGAUUUAUAGGGCAGAGUUGAAAAGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCAC- .(((...((((((........(((((((((.((((((((((.....)))))))..)))..)))))))))......))))))..))).........- ( -18.94, z-score = -2.94, R) >consensus GCGGCUAUUUAUUAUUAAAAGUGAUUUAUAGGGCAGAGUUGAAAAGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCAC_ .....................(((((((((......((((((((......))))))))..)))))))))........................... ( -8.21 = -9.09 + 0.88)

| Location | 8,115,942 – 8,116,038 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Shannon entropy | 0.26317 |
| G+C content | 0.30800 |
| Mean single sequence MFE | -15.36 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.21 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8115942 96 + 23011544 ACAAAUAAAGACCGAAAAACC---ACAAAAAAAAA-----ACAAAUAUUUUAAA-GCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .....................---...........-----..............-....((((((((..((((((.........(....))))))))))))))). ( -15.60, z-score = -2.20, R) >droGri2.scaffold_15252 13739866 102 + 17193109 ---UGUAACAUUUAAUAAAAACGGAAACAACAAAAAACCAAAAAAUAUUUUAAAAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUUAGCGCC ---...................(....)...............................(((((.((((.(((((.........(....))))))))))))))). ( -14.70, z-score = -0.42, R) >droAna3.scaffold_12916 11839379 90 - 16180835 ----AUAAAAUCUGAGAAAAA---ACCCAAAAAAAAA-------AUAUUUUAAA-GCAAGCGCUUGGAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC ----.................---.............-------..........-....((((((((..((((((.........(....))))))))))))))). ( -14.80, z-score = -1.15, R) >droEre2.scaffold_4929 17021679 90 + 26641161 ACAAAUAAAGACCGAAAAACC---ACAAAAAAAA-----------UAUUUUAAA-GCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .....................---..........-----------.........-....((((((((..((((((.........(....))))))))))))))). ( -15.60, z-score = -2.22, R) >droYak2.chr2L 10746802 101 + 22324452 ACAAAUAAAGACCGAAAAACC---ACAAAAAAAAAAAUCAACAAAUAUUUUAAA-GCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .....................---..............................-....((((((((..((((((.........(....))))))))))))))). ( -15.60, z-score = -2.18, R) >droSec1.super_3 3612317 99 + 7220098 ACAAAUAAAGACCGAAAAACC---ACAAAAAAAAAAA--AACAAAUAUUUUAAA-GCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .....................---.............--...............-....((((((((..((((((.........(....))))))))))))))). ( -15.60, z-score = -2.21, R) >droSim1.chr2L 7895455 98 + 22036055 ACAAAUAAAGACCGAAAAACC---ACAAAAAAAAAAA---ACAAAUAUUUUAAA-GCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .....................---.............---..............-....((((((((..((((((.........(....))))))))))))))). ( -15.60, z-score = -2.19, R) >consensus ACAAAUAAAGACCGAAAAACC___ACAAAAAAAAAAA___ACAAAUAUUUUAAA_GCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC ...........................................................((((((((..((((((.........(....))))))))))))))). (-15.19 = -15.21 + 0.02)

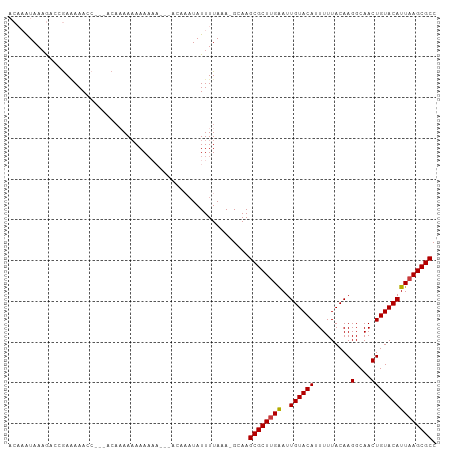
| Location | 8,115,965 – 8,116,062 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.73 |
| Shannon entropy | 0.14493 |
| G+C content | 0.29003 |
| Mean single sequence MFE | -18.35 |
| Consensus MFE | -18.31 |
| Energy contribution | -18.17 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920215 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 8115965 97 + 23011544 -----AAAAAAAAAACAAAUAUUUUAAAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCCCACGGCAUUAAGUAAAAAAUAAAA- -----.................(((((.((..((((((((..((((((.........(....))))))))))))))).....)).)))))............- ( -18.30, z-score = -1.34, R) >droAna3.scaffold_12916 11839400 94 - 16180835 ---------AAAAAAAAAAUAUUUUAAAGCAAGCGCUUGGAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCCCACGGCAUUAAGUAAAAAAAAAAAA ---------.............(((((.((..((((((((..((((((.........(....))))))))))))))).....)).)))))............. ( -17.50, z-score = -0.81, R) >droEre2.scaffold_4929 17021702 91 + 26641161 -----------AAAAAAAAUAUUUUAAAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCCCACGGCAUUAAGUAAAAAAUAAAA- -----------...........(((((.((..((((((((..((((((.........(....))))))))))))))).....)).)))))............- ( -18.30, z-score = -1.30, R) >droYak2.chr2L 10746825 103 + 22324452 AAAAAAAAAAAUCAACAAAUAUUUUAAAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCCCACGGCAUUAAGUAAAAAAAUGAAA ...........(((........(((((.((..((((((((..((((((.........(....))))))))))))))).....)).)))))........))).. ( -19.39, z-score = -1.58, R) >droSec1.super_3 3612341 99 + 7220098 ---AAAAAAAAAAAACAAAUAUUUUAAAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCCCACGGCAUUAAGUAAAAAAUAAAA- ---...................(((((.((..((((((((..((((((.........(....))))))))))))))).....)).)))))............- ( -18.30, z-score = -1.41, R) >droSim1.chr2L 7895478 99 + 22036055 ---AAAAAAAAAAAACAAAUAUUUUAAAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCCCACGGCAUUAAGUAAAAAAUAAAA- ---...................(((((.((..((((((((..((((((.........(....))))))))))))))).....)).)))))............- ( -18.30, z-score = -1.41, R) >consensus _____AAAAAAAAAACAAAUAUUUUAAAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCCCACGGCAUUAAGUAAAAAAUAAAA_ ......................(((((.((..((((((((..((((((.........(....))))))))))))))).....)).)))))............. (-18.31 = -18.17 + -0.14)

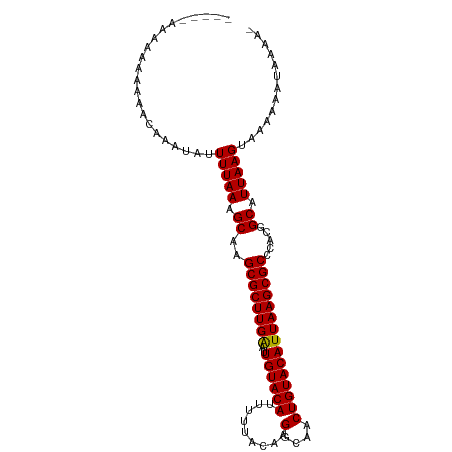
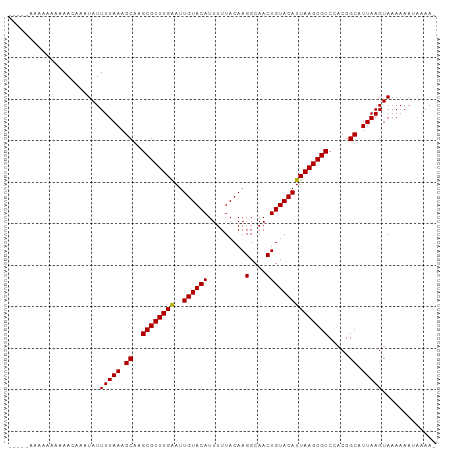
| Location | 8,115,965 – 8,116,062 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.73 |
| Shannon entropy | 0.14493 |
| G+C content | 0.29003 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987490 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 8115965 97 - 23011544 -UUUUAUUUUUUACUUAAUGCCGUGGGCGCUUAAUGUACAGUUGCCUUGUAAAAAUGUACAAUUCAAGCGCUUGCUUUAAAAUAUUUGUUUUUUUUUU----- -.....................(..(((((((((((((((.((((...))))...))))).))).)))))))..).......................----- ( -22.10, z-score = -1.99, R) >droAna3.scaffold_12916 11839400 94 + 16180835 UUUUUUUUUUUUACUUAAUGCCGUGGGCGCUUAAUGUACAGUUGCCUUGUAAAAAUGUACAAUCCAAGCGCUUGCUUUAAAAUAUUUUUUUUUU--------- ......................(..(((((((..((((((.((((...))))...))))))....)))))))..)...................--------- ( -22.00, z-score = -2.26, R) >droEre2.scaffold_4929 17021702 91 - 26641161 -UUUUAUUUUUUACUUAAUGCCGUGGGCGCUUAAUGUACAGUUGCCUUGUAAAAAUGUACAAUUCAAGCGCUUGCUUUAAAAUAUUUUUUUU----------- -.....................(..(((((((((((((((.((((...))))...))))).))).)))))))..).................----------- ( -22.10, z-score = -2.21, R) >droYak2.chr2L 10746825 103 - 22324452 UUUCAUUUUUUUACUUAAUGCCGUGGGCGCUUAAUGUACAGUUGCCUUGUAAAAAUGUACAAUUCAAGCGCUUGCUUUAAAAUAUUUGUUGAUUUUUUUUUUU ......................(..(((((((((((((((.((((...))))...))))).))).)))))))..)............................ ( -22.10, z-score = -1.70, R) >droSec1.super_3 3612341 99 - 7220098 -UUUUAUUUUUUACUUAAUGCCGUGGGCGCUUAAUGUACAGUUGCCUUGUAAAAAUGUACAAUUCAAGCGCUUGCUUUAAAAUAUUUGUUUUUUUUUUUU--- -.....................(..(((((((((((((((.((((...))))...))))).))).)))))))..).........................--- ( -22.10, z-score = -2.06, R) >droSim1.chr2L 7895478 99 - 22036055 -UUUUAUUUUUUACUUAAUGCCGUGGGCGCUUAAUGUACAGUUGCCUUGUAAAAAUGUACAAUUCAAGCGCUUGCUUUAAAAUAUUUGUUUUUUUUUUUU--- -.....................(..(((((((((((((((.((((...))))...))))).))).)))))))..).........................--- ( -22.10, z-score = -2.06, R) >consensus _UUUUAUUUUUUACUUAAUGCCGUGGGCGCUUAAUGUACAGUUGCCUUGUAAAAAUGUACAAUUCAAGCGCUUGCUUUAAAAUAUUUGUUUUUUUUUU_____ ......................(..(((((((..((((((.((((...))))...))))))....)))))))..)............................ (-22.00 = -22.00 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:12 2011