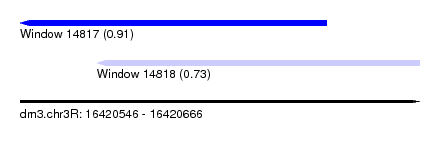
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,420,546 – 16,420,666 |
| Length | 120 |
| Max. P | 0.913362 |

| Location | 16,420,546 – 16,420,638 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.22 |
| Shannon entropy | 0.64980 |
| G+C content | 0.54004 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -12.63 |
| Energy contribution | -14.33 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
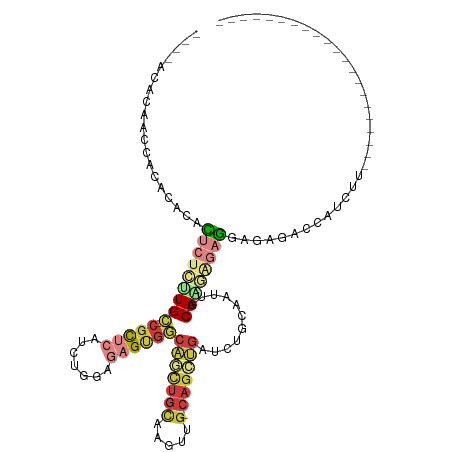
>dm3.chr3R 16420546 92 - 27905053 ----ACACAACCACACACACUCUAUUGC-CGCUCAUCUGGAGAGUGGCAGCUGCAUGUUG-CAGCUGAUCUGCAAUUGCAAGAGAGG-GAGACCAUCUU--------------------- ----...............((((.((((-(((((.......))))))))).((((.((((-(((.....))))))))))))))).((-....)).....--------------------- ( -34.50, z-score = -2.68, R) >droEre2.scaffold_4770 12494288 89 + 17746568 -------CAAACACACACACUCUCUUGC-CGCUCAUCUGGAGAGUGGCAGCUGCAAGUUG-CAGCUGAUCUCCAAUUCCA-GAGAGAAGAGACCAUCUU--------------------- -------.............((((((..-..(((.(((((((((...(((((((.....)-))))))..)))....))))-))))))))))).......--------------------- ( -32.30, z-score = -2.78, R) >droYak2.chr3R 5208651 90 - 28832112 -------CAACCACAUACACUCUCUUGC-CGCUCAUCUGGAGAGUGGCAGCUGCAAGUUG-CAGCUGAUCUCCAAUUCCAGGAGAGAAGAGACCAUCUU--------------------- -------.............((((((((-(((((.......)))))))((((((.....)-)))))..(((((.......))))).)))))).......--------------------- ( -34.20, z-score = -3.07, R) >droSec1.super_27 483552 93 + 799419 ----ACACACCCACACACACUCCCUUGC-CGCUCAUCUGGAGAGUGGCAACUGCAAGUUG-CAGCUGAUCUGCAAUUGCAAGAGAGGAGAGACCAUCUU--------------------- ----...............(((((((((-(((((.......)))))))...(((((.(((-(((.....))))))))))).))).))))..........--------------------- ( -32.30, z-score = -2.45, R) >droSim1.chr3R 22469232 93 + 27517382 ----ACACAACUACACACACUCUCUUGC-CGCUCAUCUGGAGAGUGGCAGUUGCAAGUUG-CAGCUGAUCUGCAAUUGCAAGAGAGGAGAGACCAUCUU--------------------- ----...............(((((((((-(((((.......)))))))))((((((.(((-(((.....)))))))))))))))))((((.....))))--------------------- ( -35.10, z-score = -2.70, R) >droPer1.super_9 1351024 120 - 3637205 GCGUCAUUAGGCUGGCUGCUGCUGCUGCUCGUGGGGCUGAGGGGGGAGUACUGUGGAGUGCCAGUGGAGUGGCUGUGCCAGUGCAAGUGGAAGUGGCAUUGACGCUGCCGCUGCCGUUUC (((.......((((((.((.((..((.(.(.(((.(((..(.((......)).)..))).)))).).))..)).))))))))(((.((((.((((.......)))).))))))))))... ( -46.60, z-score = 1.06, R) >droMoj3.scaffold_6500 29540952 92 + 32352404 ----CCUCAAGCUGACCCAGGAGGCUGU-CGCCGAUCUGGAGCGCAACAAGAAGGAGCUG-GAGCAGACCAUCCAGCGCAAG-GACAAGGAACUGUCCU--------------------- ----......(((...(((((.(((...-.)))..))))))))((..(.....)..((((-((........)))))))).((-((((......))))))--------------------- ( -31.60, z-score = -0.60, R) >consensus ____ACACAACCACACACACUCUCUUGC_CGCUCAUCUGGAGAGUGGCAGCUGCAAGUUG_CAGCUGAUCUGCAAUUGCAAGAGAGGAGAGACCAUCUU_____________________ ...................((((((((..(((((.......))))).((((((........))))))...........)))))))).................................. (-12.63 = -14.33 + 1.70)

| Location | 16,420,569 – 16,420,666 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 57.94 |
| Shannon entropy | 0.76496 |
| G+C content | 0.51474 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -10.34 |
| Energy contribution | -10.84 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
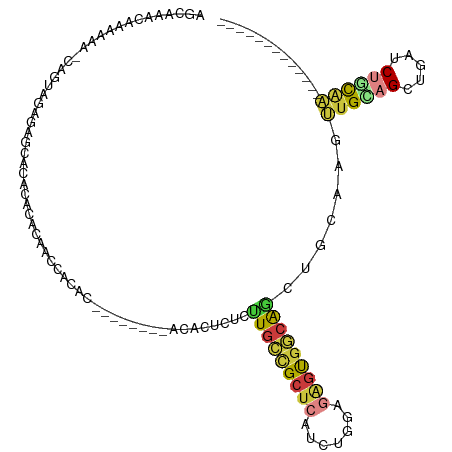
>dm3.chr3R 16420569 97 - 27905053 AGCAAACAAAACA-CAGUAGAGAGCACACACACAACCACAC--------ACACUCUAUUGCCGCUCAUCUGGAGAGUGGCAGCUGCAUGUUGCAGCUGAUCUGCAA------------- .(((.........-((((((((...................--------...))))))))((((((.......))))))(((((((.....)))))))...)))..------------- ( -30.05, z-score = -1.72, R) >droEre2.scaffold_4770 12494311 89 + 17746568 AGCAAACAAAAAG-CAGUAGAGAGCA--------AACACAC--------ACACUCUCUUGCCGCUCAUCUGGAGAGUGGCAGCUGCAAGUUGCAGCUGAUCUCCAA------------- ...........((-(.((((((((..--------.......--------...))))).))).)))....((((((....(((((((.....))))))).)))))).------------- ( -32.50, z-score = -2.58, R) >droYak2.chr3R 5208675 76 - 28832112 ----------------------AGCAAACACACAACCACAU--------ACACUCUCUUGCCGCUCAUCUGGAGAGUGGCAGCUGCAAGUUGCAGCUGAUCUCCAA------------- ----------------------...................--------.((((((((............)))))))).(((((((.....)))))))........------------- ( -24.00, z-score = -1.93, R) >droSec1.super_27 483576 98 + 799419 AGCAAACAAAAAAAAAGUAGAGAGCACACACACACCCACAC--------ACACUCCCUUGCCGCUCAUCUGGAGAGUGGCAACUGCAAGUUGCAGCUGAUCUGCAA------------- ................(((((.(((................--------........(((((((((.......))))))))).(((.....))))))..)))))..------------- ( -26.30, z-score = -1.40, R) >droSim1.chr3R 22469256 96 + 27517382 AGCAAACAAAAAA--AGUAGAGAGCACACACACAACUACAC--------ACACUCUCUUGCCGCUCAUCUGGAGAGUGGCAGUUGCAAGUUGCAGCUGAUCUGCAA------------- .............--.(((((....................--------.((((((((............)))))))).(((((((.....))))))).)))))..------------- ( -27.50, z-score = -1.00, R) >droPer1.super_9 1351064 119 - 3637205 AGCGAACUAAACACAAACUGAAUGCAAAAUUUGCAGCAUGCGUCAUUAGGCUGGCUGCUGCUGCUGCUCGUGGGGCUGAGGGGGGAGUACUGUGGAGUGCCAGUGGAGUGGCUGUGCCA ......................((((.....))))(((((.((((((..((((((..(..(((((.(((.(........).))).))))..)..)...))))))..))))))))))).. ( -36.20, z-score = 1.94, R) >droMoj3.scaffold_6500 29540975 97 + 32352404 AGAAGUCCAAGCG-CAAGGUUGAGGGUGACCUCAAGCUGAC--------CCAGGAGGCUGUCGCCGAUCUGGAGCGCAACAAGAAGGAGCUGGAGCAGACCAUCCA------------- ..........(((-(.((.((((((....)))))).))...--------(((((.(((....)))..))))).))))........(((..(((......)))))).------------- ( -38.60, z-score = -2.06, R) >consensus AGCAAACAAAAAA_CAGUAGAGAGCACACACACAACCACAC________ACACUCUCUUGCCGCUCAUCUGGAGAGUGGCAGCUGCAAGUUGCAGCUGAUCUGCAA_____________ .........................................................(((((((((.......))))))))).......((((((.....))))))............. (-10.34 = -10.84 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:23 2011