| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,414,529 – 16,414,637 |
| Length | 108 |
| Max. P | 0.783307 |

| Location | 16,414,529 – 16,414,637 |
|---|---|
| Length | 108 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 66.51 |
| Shannon entropy | 0.76281 |
| G+C content | 0.47968 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -10.81 |
| Energy contribution | -10.24 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
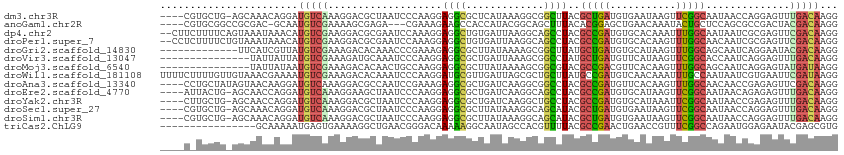
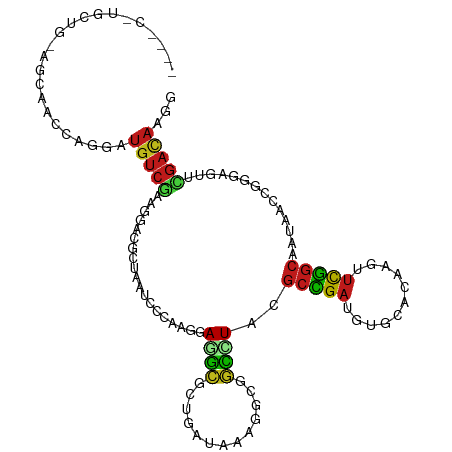
>dm3.chr3R 16414529 108 + 27905053 ----CGUGCUG-AGCAAACAGGAUGUCAAAGGACGCUAAUCCCAAGGAGGCGCUCAUAAAGGCGGCUUACGCUGAUGUGAAUAAGUUCGGCAAUAACCAGGAGUUUGACAAGG ----.((.(((-......))).))(((((((((((((..(((...))))))..(((((..((((.....))))..)))))....))))((......)).....)))))).... ( -28.10, z-score = -0.16, R) >anoGam1.chr2R 50045926 105 - 62725911 ----CGUGCGGCCGCGAC-GCAAUGUCGAAAAGCGAGA---CGAAAGAAGCCACCAUACGGCAGCUUUACACGGAGCUGAACAAAUACUGCUCCAGCGCCGACUACGACAAGG ----((((((((.(((((-.....)))(.(((((....---(....)..(((.......))).))))).)..(((((............))))).)))))).).)))...... ( -33.10, z-score = -2.02, R) >dp4.chr2 28195601 111 - 30794189 --CUUCUUUUCAGUAAAUAAACAUGUCGAAGGACGCGAAUCCAAAGGAGGCUGUGAUUAAGGCAGCCUACGCCGAUGUGCACAAAUUUGGCAAUAAUCGCGAGUUCGACAAGG --...................(.(((((((...(((((.......(.(((((((.......))))))).)(((((...........))))).....)))))..))))))).). ( -33.30, z-score = -2.28, R) >droPer1.super_7 1675811 111 - 4445127 --CCUCUUUUCUGUAAAUAAACAUGUCGAAGGACGCGAAUCCAAAGGAGGCUGUGAUUAAGGCAGCCUACGCCGAUGUGCACAAGUUUGGCAACAAUCGCGAGUUCGACAAGG --(((......(((......)))(((((((...(((((.......(.(((((((.......))))))).)(((((...........))))).....)))))..)))))))))) ( -33.50, z-score = -1.68, R) >droGri2.scaffold_14830 832953 100 + 6267026 -------------UUCAUCGUUAUGUCGAAAGACACAAACCCGAAAGAGGCGCUUAUAAAAGCGGCUUAUGCUGAUGUGCAUAAGUUUGGCAGCAAUCAGGAAUACGACAAGG -------------....((((..((((....))))....((.((..(((.(((((....))))).))).(((((....((....))....))))).)).))...))))..... ( -29.20, z-score = -2.67, R) >droVir3.scaffold_13047 6903945 98 - 19223366 ---------------UAUUAUUAUGUCGAAAGAUGCAAAUCCCAAGGAGGCGCUGAUUAAAGCGGCCUAUGCUGAUGUUCAUAAGUUCGGCACCAAUCAGGAGUUUGACAAGG ---------------.........(((....))).((((..((....(((((((......))).)))).((((((...........)))))).......))..))))...... ( -25.90, z-score = -1.21, R) >droMoj3.scaffold_6540 29489506 98 + 34148556 ---------------UAUUAUAAUGUCGAAAGACACAACUGCCAAGGAGGCGCUUAUAAAAGCGGCGUACGCCGACGUUCACAAGUUUGGCAGCAAUCAGGAGUAUGAUAAGG ---------------(((((((.((((....))))...((((((((..(.(((((....))))).)(.(((....))).).....))))))))..........)))))))... ( -29.90, z-score = -3.13, R) >droWil1.scaffold_181108 2234446 113 - 4707319 UUUUCUUUUGUUGUAAACGAAAAUGUCGAAAGACACAAAUCCCAAGGAUGCGUUGAUUAGCGCUGCUUAUGCCGAUGUCAACAAAUUUGCCAAUAAUCGUGAAUUCGAUAAGG ......(((((((...((((...((((....))))(((((((...))))..((((((...((..((....))))..))))))....))).......)))).....))))))). ( -25.10, z-score = -1.03, R) >droAna3.scaffold_13340 10533724 109 - 23697760 ----CCUGCUAUAGUAACAAGGAUGUCAAAGGACGCCAAUCCGAAAGAGGCGCUGAUCAAGGCGGCCUACGCCGAUGUUCACAAGUUUGGCAACAACCGAGAGUUCGACAAGG ----(((......(.(((..(((((((...((.((((....(....).))))))......((((.....)))))))))))....(.(((....))).)....))))....))) ( -27.90, z-score = -0.06, R) >droEre2.scaffold_4770 12488312 108 - 17746568 ----AUUACUG-AGCAACCAGGAUGUCAAAGGAAGCUAAUCCCAAGGAGGCGCUGAUCAAGGCAGCCUACGCCGAUGUGCAUAAGUUCGGCAAUAACAGAGAGUUUGACAAGG ----....(((-......)))..((((((((((......)))...(.(((((((......))).)))).)(((((...........)))))............)))))))... ( -27.50, z-score = -0.38, R) >droYak2.chr3R 5202740 108 + 28832112 ----CUUGCUG-AGCAACCAGGAUGUCAAAGGACGCUAAUCCCAAGGAGGCGCUGAUCAAGGCUGCCUACGCCGAUGUGCAUAAAUUCGGCAAUAACCGAGAGUUUGACAAGG ----((((..(-(((.....(((((((....)))....))))...(((((((((......))).))))..(((((...........))))).....))....))))..)))). ( -31.00, z-score = -0.64, R) >droSec1.super_27 477527 108 - 799419 ----CGUGCUG-AGCAAACAGGAUGUCAAAGGACGCUAAUCCCAAGGAGGCGCUUAUAAAGGCAGCAUACGCUGAUGUGAAUAAGUUCGGCAAUAACCAGGAGUUUGACAAGG ----..(((((-(((..............(((.((((..(((...)))))))))).......((((....))))..........))))))))..................... ( -24.00, z-score = 0.55, R) >droSim1.chr3R 22463013 108 - 27517382 ----CGUGCUG-AGCAAACAGGAUGUCAAAGGACGCUAAUCCCAAGGAGGCGCUUAUAAAGGCAGCAUACGCUGAUGUGAAUAAGUUCGGCAAUAACCAGGAGUUUGACAAGG ----..(((((-(((..............(((.((((..(((...)))))))))).......((((....))))..........))))))))..................... ( -24.00, z-score = 0.55, R) >triCas2.ChLG9 12804801 97 - 15222296 ----------------GCAAAAAUGAGUGAAAAGGCUGAACGGGACAAAAAGGCAAUAGCCACGUUUUACGCCGAACUGAACCGUUUCGGCCAGAAUGGAGAAUACGAGCGUG ----------------((...............((((((((((..((....(((....)))..((((......))))))..))).)))))))....((.......)).))... ( -23.90, z-score = -1.03, R) >consensus ____C_UGCUG_AGCAACCAGGAUGUCGAAGGACGCUAAUCCCAAGGAGGCGCUGAUAAAGGCGGCCUACGCCGAUGUGCACAAGUUCGGCAAUAACCGGGAGUUCGACAAGG .......................(((((...................((((.............))))..(((((...........)))))..............)))))... (-10.81 = -10.24 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:21 2011