
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,397,672 – 16,397,774 |
| Length | 102 |
| Max. P | 0.631069 |

| Location | 16,397,672 – 16,397,774 |
|---|---|
| Length | 102 |
| Sequences | 14 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.98 |
| Shannon entropy | 0.47461 |
| G+C content | 0.55732 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -12.76 |
| Energy contribution | -13.61 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16397672 102 + 27905053 ---AUCUACCACAAGACGGAGGAGGGCGAGCAGCCCAAGACCGUGUGCAAGGAGACGGUCAAGUACUUCUUCGGAUGCUCGGACAGCAU-CCUGCCGUCGGUGAUC---- ---......(((..(((((((((((((.....))))..((((((.(......).)))))).......)))))(((((((.....)))))-))...)))).)))...---- ( -42.10, z-score = -2.57, R) >droSim1.chr3R 22445990 102 - 27517382 ---AUCUACCACAAGACGGAGGAGGGCGAGGAGCCCAAGACCGUGUGCAAGGAGACGGUCAAGUACUUCUUCGGAUGCUCGGACAGCAU-CCUGCCGUCGGUGAUC---- ---......(((..(((((((((((((.....))))..((((((.(......).)))))).......)))))(((((((.....)))))-))...)))).)))...---- ( -42.10, z-score = -2.43, R) >droSec1.super_27 460566 102 - 799419 ---AUCUACCACAAGACGGAGGAGGGCGAGGAGCCCAAGACCGUGUGCAAGGAGACGGUCAAGUACUUCUUCGGAUGCUCGGACAGCAU-CCUGCCGUCGGUGAUC---- ---......(((..(((((((((((((.....))))..((((((.(......).)))))).......)))))(((((((.....)))))-))...)))).)))...---- ( -42.10, z-score = -2.43, R) >droYak2.chr3R 5183500 102 + 28832112 ---AUUUACCACAAGACGGAGGAGGGCGAGCAGCCCAAGACCGUGUGCAAGGAGACGGUGAAGUACUUCUUCGGAUGCUCGGACAGCAU-CCUUCCGUCGGUGAUC---- ---..(((((....(((((((..((((.....))))...(((((.(......).)))))((((.....))))(((((((.....)))))-))))))))))))))..---- ( -41.10, z-score = -2.77, R) >droEre2.scaffold_4770 12471376 102 - 17746568 ---AUCUACCACAAGACGGAGGAGGGCGAGCAGCCCAAGACCGUGUGCAAGGAGACGGUCAAGUACUUCUUCGGAUGCUCGGACAGCAU-CCUUCCGUCGGUGAUC---- ---......(((..(((((((((((((.....))))..((((((.(......).))))))..........))(((((((.....)))))-))))))))).)))...---- ( -42.10, z-score = -2.79, R) >droAna3.scaffold_13340 10518653 102 - 23697760 ---AUCUACCACAAGACGGAGGAGGGCGAACAGCCCAAGACCGUCUGCAAGGAGACGGUCAAGUACUUCUUUGGCUGCUCGGACAGCAU-CCUGCCGUCGGUGAUA---- ---......(((..(((((((((((((.....))))..((((((((......)))))))).............((((......)))).)-))).))))).)))...---- ( -44.00, z-score = -3.41, R) >dp4.chr2 28178896 102 - 30794189 ---AUUUACCACAAGACCGAGGAGGGGGAGCAGCCCAAGACCGUGUGCAAGGAGACGGUCAAGUACUUCUUCGGCUGCUCGGACAGCAU-UCUGCCGUCGGUGAUA---- ---..(((((...((.((((((((((((.....)))..((((((.(......).)))))).....))))))))))).....(((.((..-...)).))))))))..---- ( -40.40, z-score = -2.30, R) >droPer1.super_7 1659016 102 - 4445127 ---AUUUACCACAAGACCGAGGAGGGGGAGCAGCCCAAGACCGUGUGCAAGGAGACGGUCAAGUACUUCUUCGGCUGCUCGGACAGCAU-UCUGCCGUCGGUGAUA---- ---..(((((...((.((((((((((((.....)))..((((((.(......).)))))).....))))))))))).....(((.((..-...)).))))))))..---- ( -40.40, z-score = -2.30, R) >droWil1.scaffold_181108 2214567 102 - 4707319 ---AUUUACCACAAGACCGAGGAGGGCGAGGAGCCCAAAACGGUGUGCAAGGAGACAGUCAAAUAUUUCUUUGGCUGCUCUGACAGUAU-UCUACCCUCGGUGAUC---- ---............(((((((.((((.....)))).....((.((((..((((.((((((((......))))))))))))....))))-.))..)))))))....---- ( -38.60, z-score = -3.45, R) >droVir3.scaffold_13047 8805167 102 - 19223366 ---AUUUACCACAAAACGGAGGAGGGCGAACAGCCAAAGACCGUCUGCAAAGAGACGGUCAAAUAUUUCUUUGGCUGCUCGGACAGCAU-ACUGCCCUCGGUCAUC---- ---..............((..((((((((.((((((((((((((((......)))))))).........)))))))).))....((...-.))))))))..))...---- ( -37.80, z-score = -3.93, R) >droGri2.scaffold_14624 4091461 102 - 4233967 ---AUUUACCACAAGACGGAGGAGGGUGAACAGCCCAAGACCGUCUGCAAGGAGACGGUCAAAUAUUUCUUUGGCUGCUCGGACAGCAU-UUUACCCUCGGUGAUA---- ---..(((((......(((((((((((.....))))..((((((((......))))))))......)))))))((((......))))..-.........)))))..---- ( -36.70, z-score = -3.02, R) >droMoj3.scaffold_6540 5595867 102 - 34148556 ---AUCUACCACAAGACCGAGGAGGGGGAGCAGCCCAAGACCGUGUGCAAGGAGACGGUCAAGUAUUUCUUUGGCUGCUCCGACAGCAU-CCUGCCCUCGGUGAUC---- ---............(((((((.((((((((((((...((((((.(......).))))))(((.....))).)))))))))(....)..-)))..)))))))....---- ( -43.70, z-score = -3.05, R) >anoGam1.chr2R 8790080 102 + 62725911 ---AUCUACAGCCCGCAGGCGAACGGCAAGCCGGACACGACGAUGAAGCGCGAGGUGUUCAAGUAUCUGCUCGGUAUGUCAAAGUCCGA-UCUGCCCUCGGUCAUG---- ---.......(.(((..(((((.(((....))((((..(((.(((..(.((.(((((......))))))).)..))))))...))))).-)).)))..))).)...---- ( -27.70, z-score = 1.35, R) >triCas2.ChLG8 14583840 110 + 15773733 UCGUUCCAGGAGCAGUUGGCUAAUUUGCAAGGGUCGAUCAACCCGUAUUGGGAGUUGGUUAAGUGGGUUUUUCGCCGGUCGGAUCACACGUUUAUCGGCAUUGUCACGGC ((((..(((...((.(((((((((((.(((((((......))))...))).))))))))))).))........((((((((.......))...)))))).)))..)))). ( -33.20, z-score = -0.97, R) >consensus ___AUCUACCACAAGACGGAGGAGGGCGAGCAGCCCAAGACCGUGUGCAAGGAGACGGUCAAGUACUUCUUCGGCUGCUCGGACAGCAU_CCUGCCGUCGGUGAUC____ ......((((......(.((((((((.......)))...(((((.(......).)))))........))))).).((((.....))))...........))))....... (-12.76 = -13.61 + 0.85)

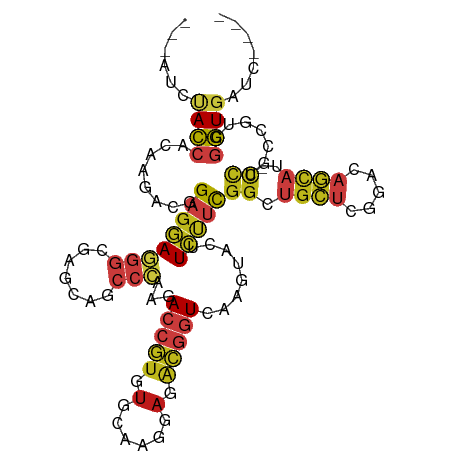

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:16 2011