| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,114,895 – 8,114,987 |
| Length | 92 |
| Max. P | 0.806292 |

| Location | 8,114,895 – 8,114,987 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.33 |
| Shannon entropy | 0.26020 |
| G+C content | 0.27398 |
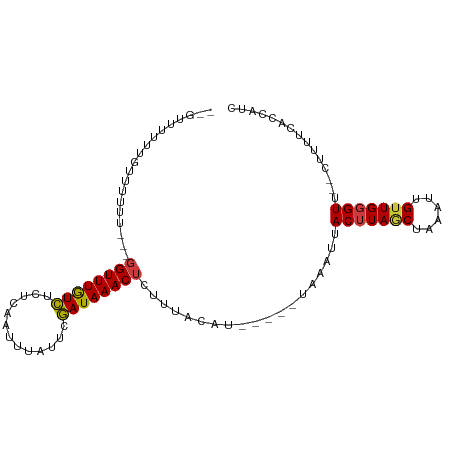
| Mean single sequence MFE | -11.82 |
| Consensus MFE | -8.95 |
| Energy contribution | -8.61 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806292 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
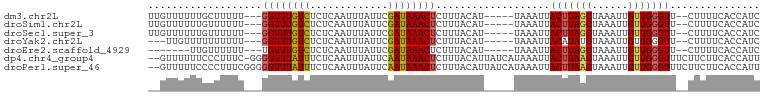
>dm3.chr2L 8114895 92 - 23011544 UUGUUUUUUGCUUUUU---GGUUUGUCUCUCAAUUUAUUCGAUAAACUCUUUACAU-----UAAAUUACUUAGCUAAAUUGUUGGGUU--CUUUUCACCAUC ..((((..((......---((((((((.............)))))))).....)).-----.)))).(((((((......))))))).--............ ( -11.32, z-score = -1.37, R) >droSim1.chr2L 7894416 92 - 22036055 UUGUUUUUUGUUUUUU---GGUUUGUCUCUCAAUUUAUUCGAUAAACUCUUUACAU-----UAAAUUACUUAGCUAAAUUGUUGGGUU--CUUUUCACCAUC ..((((..(((.....---((((((((.............))))))))....))).-----.)))).(((((((......))))))).--............ ( -13.72, z-score = -2.43, R) >droSec1.super_3 3611278 92 - 7220098 UUGUUUUUUGUUUUUU---GGUUUGUCUCUCAAUUUAUUCGAUAAACUCUUUACAU-----UAAAUUACUUAGCUAAAUUGUUGGGUU--CUUUUCACCAUC ..((((..(((.....---((((((((.............))))))))....))).-----.)))).(((((((......))))))).--............ ( -13.72, z-score = -2.43, R) >droYak2.chr2L 10745764 89 - 22324452 ---UUGUUUUUUUUUU---GGUUUGUCUCUCAAUUUAUUCGAUAAACUCUUUACAU-----UAAAUUACAUAUCUAAAUUGUUGGGUU--CUUUUCACCAUC ---............(---(((......((((((......((((............-----.........))))......))))))..--......)))).. ( -7.14, z-score = -0.07, R) >droEre2.scaffold_4929 17020652 85 - 26641161 -------UUGUUUUUU---UGUUUGUCUCUCAAUUUAUUCGAUAAACUCUUUACAU-----UAAAUUACUUAGCUAAAUUGUUGGGUU--CUUUUCACCAUC -------.(((.....---.(((((((.............))))))).....))).-----.(((..(((((((......))))))).--.)))........ ( -11.12, z-score = -2.03, R) >dp4.chr4_group4 5636638 99 + 6586962 --GUUUUUUCCCUUUC-GGGGUUUAUUUCUCAAUUUAUUCAAUAAACUCUUUACAUUAUCAUAAAUUACUUAACUAAAUUGUUGGGUUUCUUCUUCACCAUU --..............-((((((((((.............)))))))))).................(((((((......)))))))............... ( -11.52, z-score = -2.11, R) >droPer1.super_46 108525 100 - 590583 --GUUUUUCCCCUUUCGGGGGUUUAUUUCUCAAUUUAUUCAAUAAACUCUUUACAUUAUCAUAAAUUACUUAACUAAAUUGUUGGGUUUCUUCUUCACCAUU --......((((....))))(((((((.............)))))))....................(((((((......)))))))............... ( -14.22, z-score = -2.39, R) >consensus __GUUUUUUGUUUUUU___GGUUUGUCUCUCAAUUUAUUCGAUAAACUCUUUACAU_____UAAAUUACUUAGCUAAAUUGUUGGGUU__CUUUUCACCAUC ...................((((((((.............))))))))...................(((((((......)))))))............... ( -8.95 = -8.61 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:09 2011