
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,383,431 – 16,383,544 |
| Length | 113 |
| Max. P | 0.885768 |

| Location | 16,383,431 – 16,383,544 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 136 |
| Reading direction | reverse |
| Mean pairwise identity | 56.15 |
| Shannon entropy | 0.78690 |
| G+C content | 0.43484 |
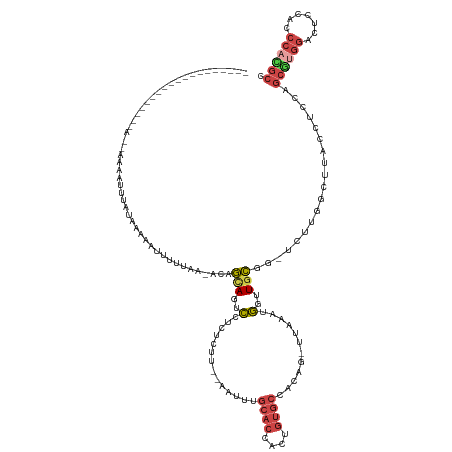
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -9.08 |
| Energy contribution | -10.14 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
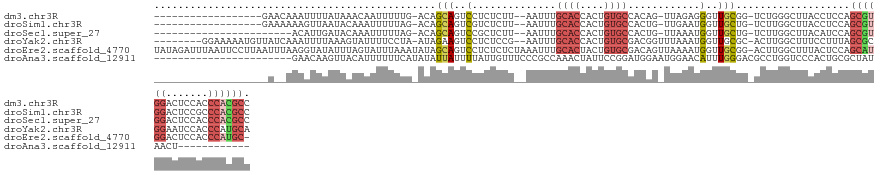
>dm3.chr3R 16383431 113 - 27905053 ------------------GAACAAAUUUUAUAAACAAUUUUUG-ACAGCAGUCCUCUCUU--AAUUUGCACCACUGUGCCACAG-UUAGAGGGUUGCGG-UCUGGGCUUACCUCCAGCGUGGACUCCACCCACGCC ------------------........................(-((.(((..(((((...--.....((((....)))).....-..)))))..))).)-))((((......))))((((((.......)))))). ( -33.76, z-score = -2.36, R) >droSim1.chr3R 22430574 113 + 27517382 ------------------GAAAAAAGUUAAUACAAAUUUUUAG-ACAGCAGUCGUCUCUU--AAUUUGCACCACUGUGCCACUG-UUGAAUGGUUGCUG-UCUUGGCUUACCUCCAGCGUGGACUCCGCCCACGCC ------------------...(((((((......)))))))((-((((((..(((...((--(((..((((....))))....)-)))))))..)))))-)))(((.......)))((((((.......)))))). ( -33.10, z-score = -2.74, R) >droSec1.super_27 446718 108 + 799419 -----------------------ACAUUGAUACAAAUUUUUAG-ACAGCAGUCCGCUCUU--AAUUUGCACCACUGUGCCACUG-UUAAAUGGUUGCUG-UCUUGGCUUACAUCCAGCGUGGACUCCACCCACGCC -----------------------..................((-(((((((.(((...((--(((..((((....))))....)-)))).)))))))))-)))(((.......)))((((((.......)))))). ( -31.00, z-score = -2.73, R) >droYak2.chr3R 5167548 124 - 28832112 --------GGAAAAAUGUUAUCAAAUUUUAAAGUAUUUUCCUA-AUAGAAGUCCUCUCCG--AAUUUGCACCACUGUGCGACGGUUUAAAUGGUUGCGC-ACUUGGCUUUCCUUUAGCGCGGAAUCCACCCAUGCA --------(((((........(((((((...((...((((...-...))))....))..)--))))))..(((.((((((((.((....)).)))))))-)..))).)))))....(((.((.......)).))). ( -26.60, z-score = -0.17, R) >droEre2.scaffold_4770 12455788 134 + 17746568 UAUAGAUUUAAUUCCUUAAUUUAAGGUAUAUUUAGUAUUUAAAUAUAGCAGUCCUCUCUCUAAAUUUGCACUACUGUGCGACAGUUAAAAUGGUUGCGG-ACUUGGCUUUACUCCAGCAUGGACUCCACCCAUGC- ..((((.((((.(((...........(((((((((...)))))))))((((.((...........((((((....))))))..........))))))))-).)))).)))).....((((((.......))))))- ( -28.70, z-score = -0.94, R) >droAna3.scaffold_12911 2689904 101 + 5364042 -----------------------GAACAAGUUACAUUUUUUCAUAUAUUAUUUUAUUGUUUCCCGCCAAACUAUUCCGGAUGGAAUGGAACAUUUGGGACGCCUGGUCCCACUGCGCUAUAACU------------ -----------------------..................................(((...(((....(((((((....)))))))......((((((.....))))))..)))....))).------------ ( -21.00, z-score = -0.86, R) >consensus ___________________A__AAAAUUUAUAAAAAUUUUUAA_ACAGCAGUCCUCUCUU__AAUUUGCACCACUGUGCCACAG_UUAAAUGGUUGCGG_UCUUGGCUUACCUCCAGCGUGGACUCCACCCACGCC ...............................................(((..(..............((((....))))............)..)))...................((((((.......)))))). ( -9.08 = -10.14 + 1.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:14 2011