| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,349,106 – 16,349,201 |
| Length | 95 |
| Max. P | 0.918229 |

| Location | 16,349,106 – 16,349,201 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 74.78 |
| Shannon entropy | 0.44443 |
| G+C content | 0.44025 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.03 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
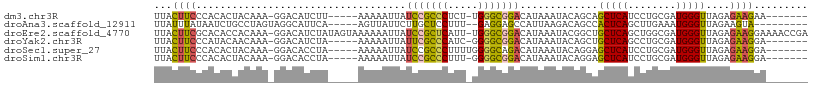
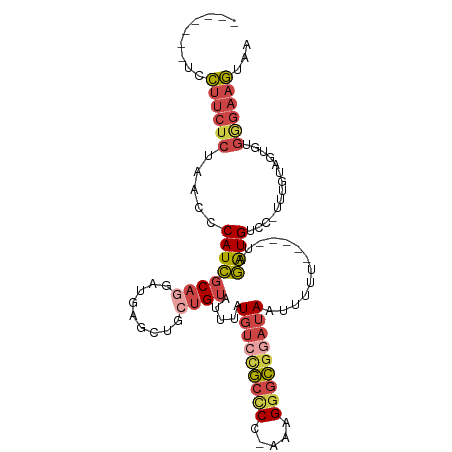
>dm3.chr3R 16349106 95 + 27905053 UUACUUCCCACACUACAAA-GGACAUCUU-----AAAAAUUAUCCGCCCUCU-UGGGCGGACAUAAAUACAGCAGCUCAUCCUGCGAUGGGUUAGAGAAGAA------- ...((((((..........-)).((((..-----........(((((((...-.)))))))..........((((......)))))))).......))))..------- ( -27.00, z-score = -2.96, R) >droAna3.scaffold_12911 2655059 93 - 5364042 UUAUUUAUAAUCUGCCUAGUAGGCAUUCA-----AGUUAUUCUUGCUCCUUU--GAGGAGCCAUUAAGACAGCCACUCAGCUUGAAAUGGGUUAGAAGUA--------- .(((((.(((((((((.....))).((((-----((((......(((((...--..)))))......((.......))))))))))..)))))).)))))--------- ( -24.40, z-score = -1.29, R) >droEre2.scaffold_4770 12421334 107 - 17746568 UUACUUCGCACACCACAAA-GGACAUCUAUAGUAAAAAAUUAUCCGCUCAUU-UGGGCGGACAUAAAUACGGCUGCUCAGCUGGCGAUGGGUUAGAGAAGGAAAACCGA ...((((.(...((.....-))..((((((.((.........(((((((...-.)))))))........(((((....))))))).))))))..).))))......... ( -27.80, z-score = -1.83, R) >droYak2.chr3R 5131650 95 + 28832112 UUACUUCCCAUACAACAAA-GGACAUCUA-----AAAAAUUAUUCGCCCAUC-GGGGCGGACAUAAAUACAGCUGCUCAGCCUGCGAUGGGUUAGAGAAGGA------- ...((((((..........-))...((((-----...........(((((((-(((((.((((..........)).)).)))).))))))))))))))))..------- ( -23.60, z-score = -1.23, R) >droSec1.super_27 412735 96 - 799419 UUACUUCCCACACUACAAA-GGACACCUA-----AAAAAUUAUCCGCCCUUUUGGGGCAGACAUAAAUACAGGAGCUCAUCCUGCGAUGGGUUAGAGAAGGA------- ...((((.(..........-.....(((.-----........((.((((.....)))).)).........)))((((((((....)))))))).).))))..------- ( -23.07, z-score = -0.95, R) >droSim1.chr3R 22396907 95 - 27517382 UUACUUCCCACACUACAAA-GGACACCUA-----AAAAAUUAUCCGCCCUUU-GGGGCGGACAUAAAUACAGGAGCUCAUCCUGCGAUGGGUUAGAGAAGGA------- ...((((.(..........-.....(((.-----........(((((((...-.))))))).........)))((((((((....)))))))).).))))..------- ( -29.17, z-score = -2.67, R) >consensus UUACUUCCCACACUACAAA_GGACAUCUA_____AAAAAUUAUCCGCCCUUU_GGGGCGGACAUAAAUACAGCAGCUCAGCCUGCGAUGGGUUAGAGAAGGA_______ ...((((...................................(((((((.....))))))).............(((((........)))))....))))......... (-15.33 = -15.03 + -0.30)
| Location | 16,349,106 – 16,349,201 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.78 |
| Shannon entropy | 0.44443 |
| G+C content | 0.44025 |
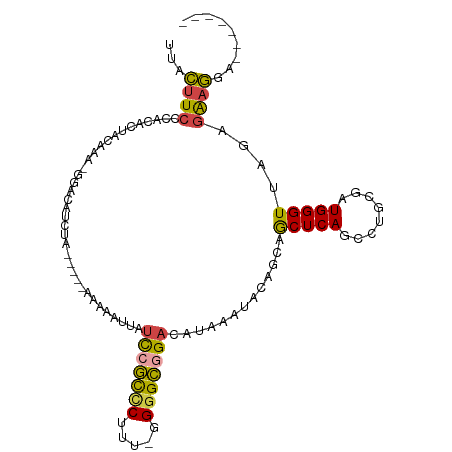
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -14.38 |
| Energy contribution | -15.66 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16349106 95 - 27905053 -------UUCUUCUCUAACCCAUCGCAGGAUGAGCUGCUGUAUUUAUGUCCGCCCA-AGAGGGCGGAUAAUUUUU-----AAGAUGUCC-UUUGUAGUGUGGGAAGUAA -------..((((((.....((((((((......))))........(((((((((.-...)))))))))......-----..))))..(-(....))...))))))... ( -30.40, z-score = -2.38, R) >droAna3.scaffold_12911 2655059 93 + 5364042 ---------UACUUCUAACCCAUUUCAAGCUGAGUGGCUGUCUUAAUGGCUCCUC--AAAGGAGCAAGAAUAACU-----UGAAUGCCUACUAGGCAGAUUAUAAAUAA ---------...((((....((((...((((....)))).....))))(((((..--...))))).)))).....-----....((((.....))))............ ( -21.50, z-score = -1.03, R) >droEre2.scaffold_4770 12421334 107 + 17746568 UCGGUUUUCCUUCUCUAACCCAUCGCCAGCUGAGCAGCCGUAUUUAUGUCCGCCCA-AAUGAGCGGAUAAUUUUUUACUAUAGAUGUCC-UUUGUGGUGUGCGAAGUAA ..((((..........))))..((((..(((....)))........(((((((.(.-...).)))))))......(((((((((.....-))))))))).))))..... ( -24.50, z-score = -0.85, R) >droYak2.chr3R 5131650 95 - 28832112 -------UCCUUCUCUAACCCAUCGCAGGCUGAGCAGCUGUAUUUAUGUCCGCCCC-GAUGGGCGAAUAAUUUUU-----UAGAUGUCC-UUUGUUGUAUGGGAAGUAA -------..((((((...(((((((..(((.(..((..........))..)))).)-))))))...(((((....-----.((.....)-)..)))))..))))))... ( -23.30, z-score = -0.45, R) >droSec1.super_27 412735 96 + 799419 -------UCCUUCUCUAACCCAUCGCAGGAUGAGCUCCUGUAUUUAUGUCUGCCCCAAAAGGGCGGAUAAUUUUU-----UAGGUGUCC-UUUGUAGUGUGGGAAGUAA -------((((.(.(((...((((((((((.....)))))).....(((((((((.....)))))))))......-----..))))...-....))).).))))..... ( -30.80, z-score = -2.47, R) >droSim1.chr3R 22396907 95 + 27517382 -------UCCUUCUCUAACCCAUCGCAGGAUGAGCUCCUGUAUUUAUGUCCGCCCC-AAAGGGCGGAUAAUUUUU-----UAGGUGUCC-UUUGUAGUGUGGGAAGUAA -------((((.(.(((...((((((((((.....)))))).....(((((((((.-...)))))))))......-----..))))...-....))).).))))..... ( -33.00, z-score = -3.20, R) >consensus _______UCCUUCUCUAACCCAUCGCAGGAUGAGCUGCUGUAUUUAUGUCCGCCCC_AAAGGGCGGAUAAUUUUU_____UAGAUGUCC_UUUGUAGUGUGGGAAGUAA .........((((((.....((((((((.........)))).....(((((((((.....))))))))).............))))..............))))))... (-14.38 = -15.66 + 1.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:10 2011