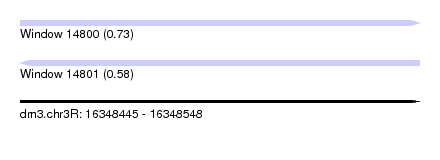
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,348,445 – 16,348,548 |
| Length | 103 |
| Max. P | 0.727092 |

| Location | 16,348,445 – 16,348,548 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.62 |
| Shannon entropy | 0.31736 |
| G+C content | 0.51579 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -22.66 |
| Energy contribution | -23.88 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16348445 103 + 27905053 ACCAUUCCUUGAGUUCCCAUCCAAGUCACCAACGUGAAGUUCGCUUUGCACAUUUGUGCUGAGCACUGAGGACCCUUGAGCAGUUUUCAGCUCUUGCCCGGAA .((..(((((.(((...........((((....)))).....((((.((((....)))).)))))))))))).....((((........))))......)).. ( -27.50, z-score = -1.07, R) >droSim1.chr3R 22396224 103 - 27517382 ACCAUUUCUUGAAUUCCCAUCCAAGUCACCAACGUGAAGUUCGCUUUGCACAUUUGUGCUGAGCACUGAGGACCCUUGAGCAGUUUUCAGCUCUUGGCCGGGA ..............((((..(((((((((....)))).....((((.((((....)))).)))).((((((((.........))))))))..)))))..)))) ( -32.10, z-score = -1.86, R) >droSec1.super_27 412061 103 - 799419 ACCAUUUCUUGAAUUCCCAUCCAAGUCACCAACGUGAAGUUCGCUUUGCACAUUUGUGCUGAGCACUGAGGGCCCUUGAGCAGUUUCCAGCUCUUGGCCGGGA ..............((((.......((((....)))).....((((.((((....)))).))))......((((...((((........))))..)))))))) ( -33.70, z-score = -1.71, R) >droYak2.chr3R 5130991 103 + 28832112 CCCCUUCCUUGAAUUUCCAUCCAAGUCACCAAUGUGAAGUUCGCUUUGCACAUUUGUGCUGAGCUCUGAGGACCCUUGAGCAGCUUUCAGCUCUUGGCCGGGA .(((.(((((((((((((((...........))).)))))))((((.((((....)))).))))...))))).((..((((........))))..))..))). ( -35.90, z-score = -2.65, R) >droEre2.scaffold_4770 12420617 103 - 17746568 CCCCUUCCUUGAAUUCCCAUCCAAGUCACCAAUGUGAAGUUCGCUUUGCACAUUUGUGCUGAGCACUGAGGGCCCUUGAGCAGCAUUCAGCUCUUGGCCGGGA .(((...((((((((..........((((....)))))))))((((.((((....)))).))))...)))((((...((((........))))..))))))). ( -34.20, z-score = -1.49, R) >droAna3.scaffold_12911 2654512 79 - 5364042 -------------CCUCCGUCCAAGCCACGACUGUGAAGUUCGCUUUGCACAUUUGGGCUAAG-------AACCCUUGAGCAGUUCU-AGCUCUUGGCAG--- -------------...........((((....((..(((....)))..)).....((((((.(-------(((.........)))))-))))).))))..--- ( -22.00, z-score = -0.64, R) >consensus ACCAUUCCUUGAAUUCCCAUCCAAGUCACCAACGUGAAGUUCGCUUUGCACAUUUGUGCUGAGCACUGAGGACCCUUGAGCAGUUUUCAGCUCUUGGCCGGGA ..............((((.(((...((((....)))).....((((.((((....)))).)))).....))).((..((((........))))..))..)))) (-22.66 = -23.88 + 1.22)
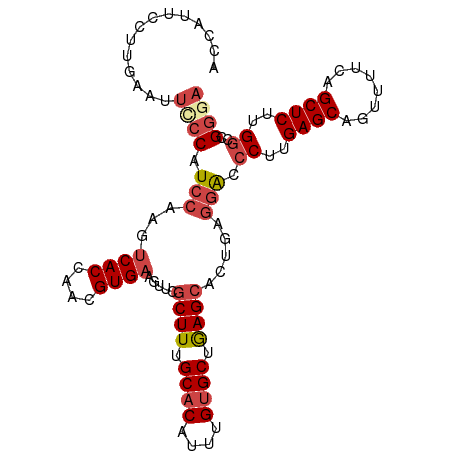

| Location | 16,348,445 – 16,348,548 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Shannon entropy | 0.31736 |
| G+C content | 0.51579 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -24.74 |
| Energy contribution | -25.93 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
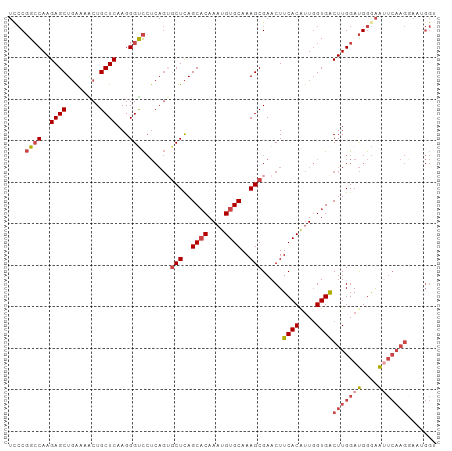
>dm3.chr3R 16348445 103 - 27905053 UUCCGGGCAAGAGCUGAAAACUGCUCAAGGGUCCUCAGUGCUCAGCACAAAUGUGCAAAGCGAACUUCACGUUGGUGACUUGGAUGGGAACUCAAGGAAUGGU ((((......((((........))))..((((.((((.((((..((((....))))..))))....((((....))))......)))).))))..)))).... ( -31.10, z-score = -0.58, R) >droSim1.chr3R 22396224 103 + 27517382 UCCCGGCCAAGAGCUGAAAACUGCUCAAGGGUCCUCAGUGCUCAGCACAAAUGUGCAAAGCGAACUUCACGUUGGUGACUUGGAUGGGAAUUCAAGAAAUGGU (((((.(((((.(((((..(((.......)))..)))))(((..((((....))))..))).....((((....))))))))).))))).............. ( -33.10, z-score = -1.39, R) >droSec1.super_27 412061 103 + 799419 UCCCGGCCAAGAGCUGGAAACUGCUCAAGGGCCCUCAGUGCUCAGCACAAAUGUGCAAAGCGAACUUCACGUUGGUGACUUGGAUGGGAAUUCAAGAAAUGGU ((((((((..((((.(....).))))...))))((.((((((..((((....))))..)))......(((....)))))).))..)))).............. ( -36.70, z-score = -1.91, R) >droYak2.chr3R 5130991 103 - 28832112 UCCCGGCCAAGAGCUGAAAGCUGCUCAAGGGUCCUCAGAGCUCAGCACAAAUGUGCAAAGCGAACUUCACAUUGGUGACUUGGAUGGAAAUUCAAGGAAGGGG .(((((((..((((........))))...)))).((...(((..((((....))))..))).....((((....))))(((((((....))))))))).))). ( -36.80, z-score = -2.00, R) >droEre2.scaffold_4770 12420617 103 + 17746568 UCCCGGCCAAGAGCUGAAUGCUGCUCAAGGGCCCUCAGUGCUCAGCACAAAUGUGCAAAGCGAACUUCACAUUGGUGACUUGGAUGGGAAUUCAAGGAAGGGG .(((((((..((((........))))...)))).((..((((..((((....))))..))))....((((....))))(((((((....))))))))).))). ( -38.00, z-score = -1.68, R) >droAna3.scaffold_12911 2654512 79 + 5364042 ---CUGCCAAGAGCU-AGAACUGCUCAAGGGUU-------CUUAGCCCAAAUGUGCAAAGCGAACUUCACAGUCGUGGCUUGGACGGAGG------------- ---(((((((((((.-......))))..((((.-------....))))......((...((((.((....)))))).)))))).)))...------------- ( -23.50, z-score = -0.35, R) >consensus UCCCGGCCAAGAGCUGAAAACUGCUCAAGGGUCCUCAGUGCUCAGCACAAAUGUGCAAAGCGAACUUCACAUUGGUGACUUGGAUGGGAAUUCAAGGAAUGGU ....((((..((((........))))...))))......(((..((((....))))..))).....((((....))))(((((((....)))))))....... (-24.74 = -25.93 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:08 2011