
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,347,323 – 16,347,439 |
| Length | 116 |
| Max. P | 0.644703 |

| Location | 16,347,323 – 16,347,385 |
|---|---|
| Length | 62 |
| Sequences | 4 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 98.39 |
| Shannon entropy | 0.02617 |
| G+C content | 0.52419 |
| Mean single sequence MFE | -12.45 |
| Consensus MFE | -12.23 |
| Energy contribution | -12.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16347323 62 + 27905053 UCCAGACAUCGCACAGCAGGCAUCCUGUUUGCAUACACACUCACCCUCGAACAGGUGCAAAA ..........(((.((((((...))))))))).........((((........))))..... ( -13.00, z-score = -1.19, R) >droYak2.chr3R 5129844 62 + 28832112 CCCAGACAUCGCACAGCAGUCAUCCUGUUUGCAUACACACUCACCCUCGAACAGGUGCAAAA ..........(((.(((((.....)))))))).........((((........))))..... ( -10.80, z-score = -0.84, R) >droSec1.super_27 410971 62 - 799419 CCCAGACAUCGCACAGCAGGCAUCCUGUUUGCAUACACACUCACCCUCGAACAGGUGCAAAA ..........(((.((((((...))))))))).........((((........))))..... ( -13.00, z-score = -1.06, R) >droSim1.chr3R 22394273 62 - 27517382 CCCAGACAUCGCACAGCAGGCAUCCUGUUUGCAUACACACUCACCCUCGAACAGGUGCAAAA ..........(((.((((((...))))))))).........((((........))))..... ( -13.00, z-score = -1.06, R) >consensus CCCAGACAUCGCACAGCAGGCAUCCUGUUUGCAUACACACUCACCCUCGAACAGGUGCAAAA ..........(((.(((((.....)))))))).........((((........))))..... (-12.23 = -12.23 + -0.00)

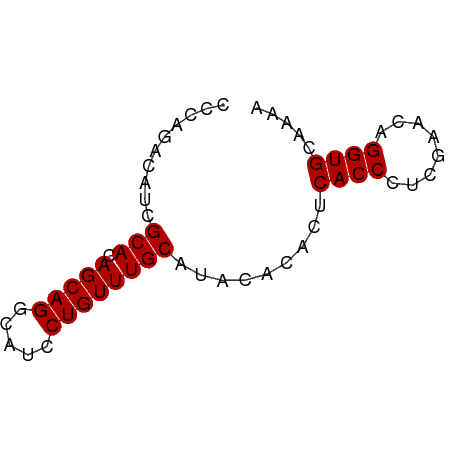

| Location | 16,347,385 – 16,347,439 |
|---|---|
| Length | 54 |
| Sequences | 4 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 88.99 |
| Shannon entropy | 0.17761 |
| G+C content | 0.40752 |
| Mean single sequence MFE | -18.70 |
| Consensus MFE | -15.49 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.644703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16347385 54 - 27905053 ----GCAAGUACGUAUGUGUGCAUAUACACAUGUAAGUAUGCGCAGGUGUAUUUGUAU ----((((((((..((.(((((((((((....))..))))))))).)))))))))).. ( -19.60, z-score = -2.01, R) >droYak2.chr3R 5129906 50 - 28832112 --------GUACGUAUGUGUGCAUAUACUCGUGUAAGUGUGCGCAGGUGUAUUUGUAU --------(((((..((((..(((.(((....))).)))..))))..)))))...... ( -17.60, z-score = -2.10, R) >droSec1.super_27 411033 54 + 799419 ----GCAAGUACGUAUGUGUGCAUAUACACGUGUAAGUAUGCGCAGGUGUAUUUGUAU ----((((((((..((.(((((((((((....))..))))))))).)))))))))).. ( -19.60, z-score = -1.95, R) >droSim1.chr3R 22394335 58 + 27517382 GCAAGUACGUACGUAUGUGUGCAUAUACACGUGUAAGUAUGCGCAGGUGUAUUUGUAU ((((((((((.((((((..(((((......)))))..))))))...)))))))))).. ( -18.00, z-score = -0.37, R) >consensus ____GCAAGUACGUAUGUGUGCAUAUACACGUGUAAGUAUGCGCAGGUGUAUUUGUAU ....((((((((..((.(((((((((((....))..))))))))).)))))))))).. (-15.49 = -16.43 + 0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:07 2011