| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,341,331 – 16,341,409 |
| Length | 78 |
| Max. P | 0.982402 |

| Location | 16,341,331 – 16,341,409 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 55.74 |
| Shannon entropy | 0.76563 |
| G+C content | 0.48073 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -8.84 |
| Energy contribution | -8.27 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
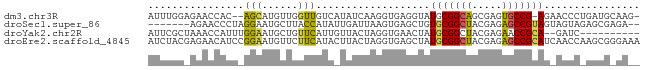
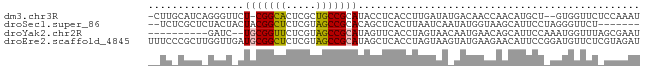
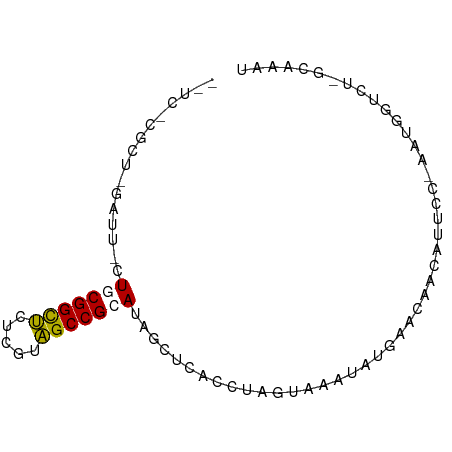
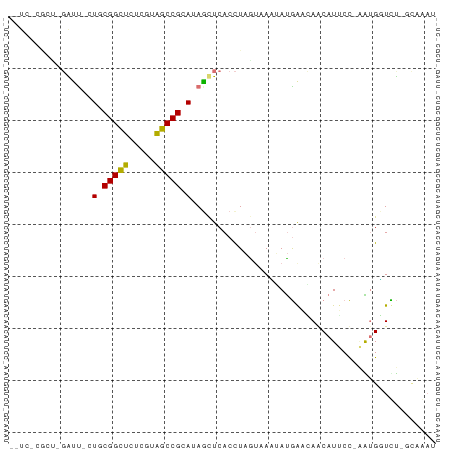
>dm3.chr3R 16341331 78 + 27905053 AUUUGGAGAACCAC--AGCAUGUUGGUUGUCAUAUCAAGGUGAGGUAUGCGGCAGCGAGUGCCG-AGAACCCUGAUGCAAG- ..(((((.(((((.--.......))))).)).(((((.(((..(((((.((....)).))))).-...))).)))))))).- ( -23.10, z-score = -1.05, R) >droSec1.super_86 62560 73 - 128402 -------AGAACCCUAGGAAUGCUUACCAUAUUGAUUAAGUGAGCUGUGCGGCUACGAGAGCCGUAGUAGUAGAGCGAGA-- -------..............(((((..........)))))..(((.(((.((((((.....)))))).))).)))....-- ( -17.50, z-score = -1.03, R) >droYak2.chr2R 205979 70 + 21139217 AUUCGCUAAACCAUUUGGAAUGCUGUUCAUUGUUACUAGGUGAACUAUGCGGCUACGAGAACCGCA--GAUC---------- ....((....((....))...)).(((((((.......)))))))..(((((.........)))))--....---------- ( -14.50, z-score = -0.39, R) >droEre2.scaffold_4845 2169216 82 + 22589142 AUCUACGAGAACAUCCGGAAUGUUCUUCAUACUUACUAGGUGAGCUAUGCGGCUACGAGAGCCGCAUCAACCAAGCGGGAAA ......((((((((.....))))))))....((..((.(((.....((((((((.....))))))))..))).))..))... ( -26.50, z-score = -2.67, R) >consensus AUUUGC_AAAACACU_GGAAUGCUGUUCAUAAUUACUAGGUGAGCUAUGCGGCUACGAGAGCCGCAG_AACC_AGCG_GA__ ................(((......)))...................(((((((.....)))))))................ ( -8.84 = -8.27 + -0.56)
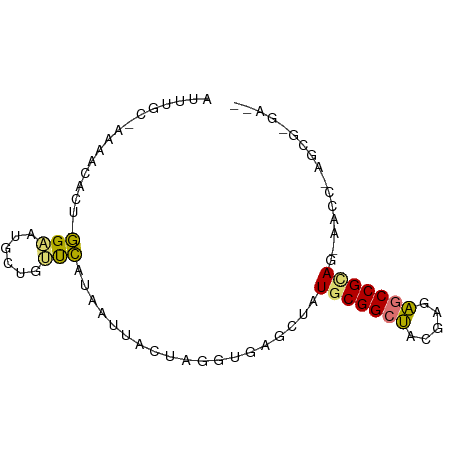
| Location | 16,341,331 – 16,341,409 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 55.74 |
| Shannon entropy | 0.76563 |
| G+C content | 0.48073 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.07 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16341331 78 - 27905053 -CUUGCAUCAGGGUUCU-CGGCACUCGCUGCCGCAUACCUCACCUUGAUAUGACAACCAACAUGCU--GUGGUUCUCCAAAU -.....((((((((...-(((((.....)))))........))))))))..((.(((((.......--.))))).))..... ( -19.00, z-score = -1.10, R) >droSec1.super_86 62560 73 + 128402 --UCUCGCUCUACUACUACGGCUCUCGUAGCCGCACAGCUCACUUAAUCAAUAUGGUAAGCAUUCCUAGGGUUCU------- --....((((((......(((((.....)))))....(((.(((..........))).))).....))))))...------- ( -15.70, z-score = -1.50, R) >droYak2.chr2R 205979 70 - 21139217 ----------GAUC--UGCGGUUCUCGUAGCCGCAUAGUUCACCUAGUAACAAUGAACAGCAUUCCAAAUGGUUUAGCGAAU ----------....--(((((((.....)))))))..(((((...........))))).(((..((....))..).)).... ( -15.00, z-score = -0.99, R) >droEre2.scaffold_4845 2169216 82 - 22589142 UUUCCCGCUUGGUUGAUGCGGCUCUCGUAGCCGCAUAGCUCACCUAGUAAGUAUGAAGAACAUUCCGGAUGUUCUCGUAGAU ......(((.(((..((((((((.....)))))))).....))).)))...(((((.(((((((...))))))))))))... ( -26.80, z-score = -2.46, R) >consensus __UC_CGCU_GAUU_CUGCGGCUCUCGUAGCCGCAUAGCUCACCUAGUAAAUAUGAACAACAUUCC_AAUGGUCU_GCAAAU ..........((((...((((((.....))))))..)))).......................................... ( -8.14 = -8.07 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:05 2011