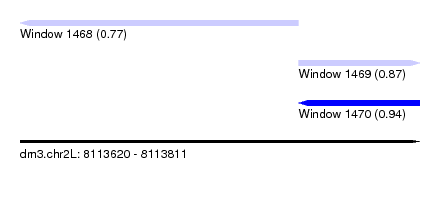
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,113,620 – 8,113,811 |
| Length | 191 |
| Max. P | 0.941834 |

| Location | 8,113,620 – 8,113,753 |
|---|---|
| Length | 133 |
| Sequences | 8 |
| Columns | 135 |
| Reading direction | reverse |
| Mean pairwise identity | 68.11 |
| Shannon entropy | 0.62028 |
| G+C content | 0.40312 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -12.43 |
| Energy contribution | -12.31 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.767680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
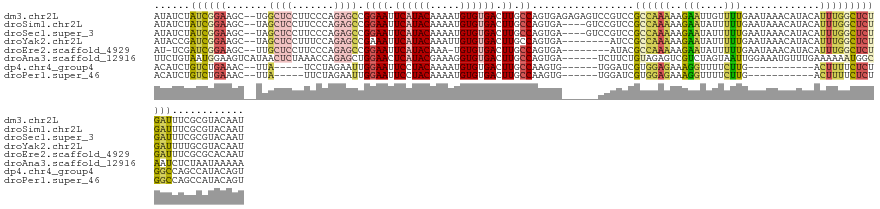
>dm3.chr2L 8113620 133 - 23011544 AUAUCUAUCGGAAGC--UGGCUCCUUCCCAGAGCCGGAAUUCAUACAAAAUGUGUGACUUGCCAGUGAGAGAGUCCGUCCGCCAAAAAGAAUUGUUUUGAAUAAACAUACAUUUGGCUCUGAUUUCGCGUACAAU .........((((((--((((((.......)))))))...((((((.....))))))))).)).(((((...(((.(...((((((......(((((.....)))))....)))))).).))))))))....... ( -33.50, z-score = -1.28, R) >droSim1.chr2L 7893137 129 - 22036055 AUAUCUAUCGGAAGC--UAGCUCCUUCCCAGAGCCGGAAUUCAUACAAAAUGUGUGACUUGCCAGUGA----GUCCGUCCGCCAAAAAGAAUAUUUUUGAAUAAACAUACAUUUGGCUCUGAUUUCGCGUACAAU .........(((((.--......)))))((((((((((....((((.....))))((((((....)))----)))..)))..((((((.....))))))...............))))))).............. ( -30.40, z-score = -1.68, R) >droSec1.super_3 3609997 129 - 7220098 AUAUCUAUCGGAAGC--UAGCUCCUUCCCAGAGCCGGAAUUCAUACAAAAUGUGUGACUUGCCAGUGA----GUCCGUCCGCCAAAAAGAAUAUUUUUGAAUAAACAUACAUUUGGCUCUGAUUUCGCGUACAAU .........(((((.--......)))))((((((((((....((((.....))))((((((....)))----)))..)))..((((((.....))))))...............))))))).............. ( -30.40, z-score = -1.68, R) >droYak2.chr2L 10744468 125 - 22324452 AUACCGAUCGGAAGC--UAGCUCCUUUCCAGAGCCGAAAUUCAUACAAAUUGUGUGACUUGCCAGUGA--------AUCCGCCAAAAAGAAUAUUUUUGAAUAAACAUACAUUUGGCUCUGAUUUUGCGUACAAU .((((((..(((.(.--...))))....((((((((((..((((((.....)))))).......(((.--------...)))((((((.....))))))............))))))))))...))).))).... ( -28.90, z-score = -2.22, R) >droEre2.scaffold_4929 17019370 123 - 26641161 AU-UCGAUCGGAAGC--UUGCUCCUUCCCAGAGCCGGAAUUCAUACAAA-UGUGUGACUUGCCAGUGA--------AUACGCCAAAAAGAAUAUUUUUGAAUAAACAUACAUUUGGCUCUGAUUUCGCGCACAAU ..-.(((..(((((.--......)))))(((((((((((.((((((...-.)))))).)).)).....--------......((((((.....))))))...............)))))))...)))........ ( -30.50, z-score = -1.84, R) >droAna3.scaffold_12916 11836963 129 + 16180835 UUCUGUAAUGGAAGUCAUAACUCUAAACCAGAGCUGGAACUCAUACGAAAGGUGUGACUUGCCAGUGA------UCUUCUGUAGAGUCGUCUAGUAAUUGGAAAUGUUUGAAAAAAUGGCAAUCUCUAAUAAAAA ((((.....))))(((((.((((((....((((((((((.((((((.....)))))).)).)))))..------)))....))))))..(((((...))))).............)))))............... ( -27.20, z-score = -0.85, R) >dp4.chr4_group4 5635212 111 + 6586962 ACAUCUGUCUGAAAC--UUA-----UCCUAGAAUUGGAAUUCCUACAAAAUGUGUGACUUGCCAAGUG------UGGAUCGUGGAGAAAGGUUUUCUUG-----------ACUUUUCUCUGGCCAGCCAUACAGU ...............--...-----........((((((.((.(((.....))).)).)).))))(((------(((...(((((((((((((.....)-----------)))))))))).))...))))))... ( -27.00, z-score = -0.54, R) >droPer1.super_46 107305 111 - 590583 ACAUCUGUCUGAAAC--UUA-----UUCUAGAAUUGGAAUUCCUACAAAAUGUGUGACUUGCCAAGUG------UGGAUCGUGGAGAAAGGUUUUCUUG-----------ACUUUUCUCUGGCCAGCCAUACAGU .......((((((..--...-----))).))).((((((.((.(((.....))).)).)).))))(((------(((...(((((((((((((.....)-----------)))))))))).))...))))))... ( -28.30, z-score = -0.99, R) >consensus AUAUCUAUCGGAAGC__UAGCUCCUUCCCAGAGCCGGAAUUCAUACAAAAUGUGUGACUUGCCAGUGA______CCGUCCGCCAAAAAGAAUAUUUUUGAAUAAACAUACAUUUGGCUCUGAUUUCGCGUACAAU ......((((((.......................((((.((((((.....)))))).)).)).................((((((.........................))))))))))))............ (-12.43 = -12.31 + -0.12)
| Location | 8,113,753 – 8,113,811 |
|---|---|
| Length | 58 |
| Sequences | 5 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 97.24 |
| Shannon entropy | 0.04979 |
| G+C content | 0.56552 |
| Mean single sequence MFE | -18.70 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
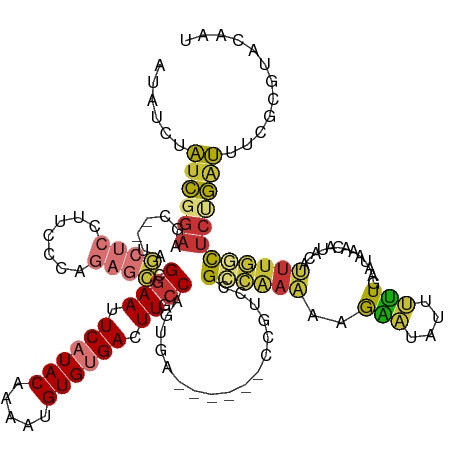
>dm3.chr2L 8113753 58 + 23011544 GGAAAUGAGUAUAUGGUGGAGGAGCCAUCAGCCGGCCAGGCUGAGAUUCCUGCGAACC ..............((((.(((((...((((((.....))))))..))))).)..))) ( -18.90, z-score = -1.43, R) >droEre2.scaffold_4929 17019493 58 + 26641161 GGAGAUGAGAAUAUGGUGGAGGAGCCAUCAGCCGGCCAGGCUGAGAUUCCUGCGAACC ..............((((.(((((...((((((.....))))))..))))).)..))) ( -18.90, z-score = -1.43, R) >droYak2.chr2L 10744593 58 + 22324452 GGAAAUGAGUAUAUGGUGGAGGAGCCAUCAGCCAGCCAGGCUGAGAUUCCUGCGAACC ..............((((.(((((...((((((.....))))))..))))).)..))) ( -18.20, z-score = -1.46, R) >droSec1.super_3 3610126 58 + 7220098 GGAAAUGAGUAUAUGGUGGAGGAGCCAUCAGCCGGCCAGGCUGAGAUUCCUGCGAAUC ................((.(((((...((((((.....))))))..))))).)).... ( -18.60, z-score = -1.30, R) >droSim1.chr2L 7893266 58 + 22036055 GGAAAUGAGUAUAUGGUGGAGGAGCCAUCAGCCGGCCAGGCUGAGAUUCCUGCGAACC ..............((((.(((((...((((((.....))))))..))))).)..))) ( -18.90, z-score = -1.43, R) >consensus GGAAAUGAGUAUAUGGUGGAGGAGCCAUCAGCCGGCCAGGCUGAGAUUCCUGCGAACC ................((.(((((...((((((.....))))))..))))).)).... (-18.46 = -18.46 + 0.00)

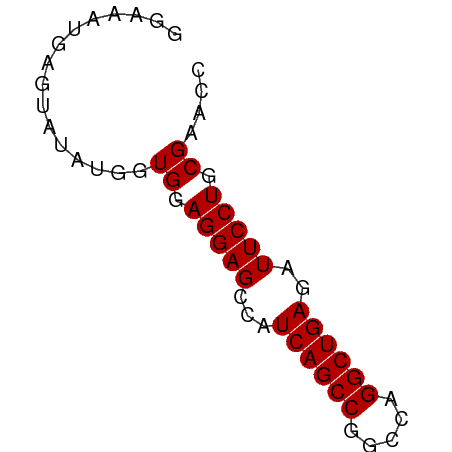
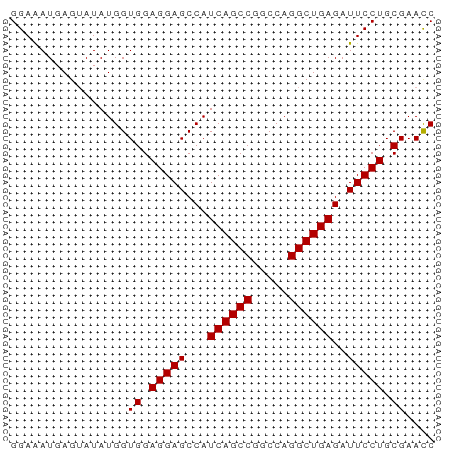
| Location | 8,113,753 – 8,113,811 |
|---|---|
| Length | 58 |
| Sequences | 5 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 97.24 |
| Shannon entropy | 0.04979 |
| G+C content | 0.56552 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.941834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8113753 58 - 23011544 GGUUCGCAGGAAUCUCAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCC (((..(.((((..(((((((.....)))))).)..))))).))).............. ( -19.10, z-score = -2.08, R) >droEre2.scaffold_4929 17019493 58 - 26641161 GGUUCGCAGGAAUCUCAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUUCUCAUCUCC (((..(.((((..(((((((.....)))))).)..))))).))).............. ( -19.10, z-score = -1.93, R) >droYak2.chr2L 10744593 58 - 22324452 GGUUCGCAGGAAUCUCAGCCUGGCUGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCC (((..(.((((..(((((((.....)))))).)..))))).))).............. ( -19.70, z-score = -2.25, R) >droSec1.super_3 3610126 58 - 7220098 GAUUCGCAGGAAUCUCAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCC .....(.((((..(((((((.....)))))).)..)))).)................. ( -16.90, z-score = -1.73, R) >droSim1.chr2L 7893266 58 - 22036055 GGUUCGCAGGAAUCUCAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCC (((..(.((((..(((((((.....)))))).)..))))).))).............. ( -19.10, z-score = -2.08, R) >consensus GGUUCGCAGGAAUCUCAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCC (((..(.((((..(((((((.....)))))).)..))))).))).............. (-17.92 = -18.12 + 0.20)

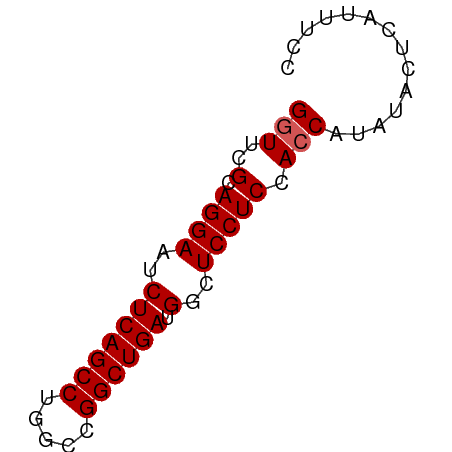
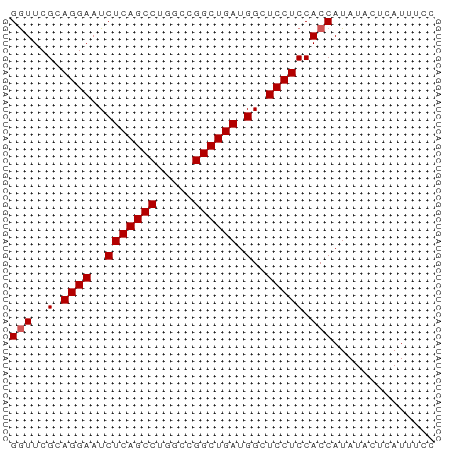
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:08 2011