| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,329,331 – 16,329,421 |
| Length | 90 |
| Max. P | 0.720100 |

| Location | 16,329,331 – 16,329,421 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 53.69 |
| Shannon entropy | 1.01218 |
| G+C content | 0.43724 |
| Mean single sequence MFE | -15.58 |
| Consensus MFE | -3.48 |
| Energy contribution | -3.91 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
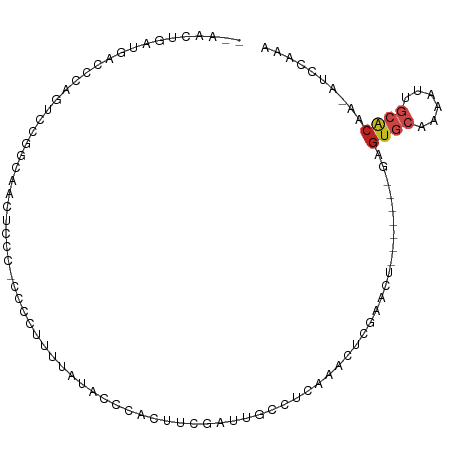
>dm3.chr3R 16329331 90 + 27905053 --AACUGAUGACCCAGUCCGGCAACUCCC-CCACUUUUAUACCCACUUCGAUUGCCUCGAACUCGAAC--------GAGUGCAAAAUUGCACAA-AUCCAAA --.((((......))))..(((((.((..-...................)))))))(((....)))..--------..(((((....)))))..-....... ( -15.00, z-score = -2.04, R) >droSim1.chr3R 22373621 96 - 27517382 --AACUGAUGACCCAGUCCGGCAACUCCC-CCACUUUUAUACCCACUUCGAUUGCCUCGAACUCGAACUCGA--ACGAGUGCAAAAUUGCACAA-AUCCAAA --.((((......))))..(((((.((..-...................)))))))(((...(((....)))--.)))(((((....)))))..-....... ( -16.40, z-score = -2.06, R) >droSec1.super_27 396490 90 - 799419 --AACUGAUGACCCAGUCCGGCAACUCCC-CCACUUUUAUACCCACUUCGAUUGCCUCGAACUCGAAC--------GAGUGCAAAAUUGCACAA-AUCCAAA --.((((......))))..(((((.((..-...................)))))))(((....)))..--------..(((((....)))))..-....... ( -15.00, z-score = -2.04, R) >droYak2.chr3R 5110148 90 + 28832112 --AACUGAUGACCCAGUCCGGCAACUACC-CCUCUUUUAUACCCACUUCGAUUGCCUCAAACUCAAGC--------GAGUGCAAAAUUGCACAA-AUCCAAA --.((((......))))..(((((.....-.....................)))))............--------..(((((....)))))..-....... ( -12.57, z-score = -1.09, R) >droEre2.scaffold_4770 12404310 96 - 17746568 --AACUGAUGACCCAGUCCGGCAACUCCC-CCUCUUUUAUACCCACUUCGAUUGCCUCGAACUCGAACUCGA--ACGAGUGCAAAAUUGCACAA-AUCCAAA --.((((......))))..(((((.((..-...................)))))))(((...(((....)))--.)))(((((....)))))..-....... ( -16.40, z-score = -2.02, R) >droAna3.scaffold_12911 2642001 71 - 5364042 ---------------CACCGACUGCUUCU--CCUAUUUAUACCGAUUUUGUUGGCCU------CAAAC--------GAGUGCAAAAUUGCACAAGUUUCGAG ---------------..(((((....((.--............))....))))).((------(((((--------..(((((....)))))..)))).))) ( -15.82, z-score = -2.16, R) >dp4.chr2 15490501 84 + 30794189 --AACUGAUGACCCAUCUCCCACGAUCCAAUCCUAUUUAUACCGAUUCAGAAAGC--------CAAACU-------GAGUGCAAAAUUGCACAGAACCCAC- --....((((...)))).............................((((.....--------....))-------))(((((....))))).........- ( -9.30, z-score = -1.04, R) >droPer1.super_0 6900493 84 - 11822988 --AACUGAUGACCCAUCUCCCACGAUCCAAUCCUAUUUAUACCGAUUCAGACAGC--------CGAACU-------GAGUGCAAAAUUGCACAGAACCCAC- --....((((...))))............................(((...(((.--------....))-------).(((((....))))).))).....- ( -9.70, z-score = -0.89, R) >droWil1.scaffold_181108 2133835 96 - 4707319 AAAACUGUUGACCUA--UCUGCAGUUCUCGCCACUUUUAUACGUAUAUCCGUAGAAGGCAAA-CGAGUA-AA--ACGAGUGCAAAAUUGCACGAUAUCCGAG ..((((((.((....--)).))))))((((....((((((.(((....((......))...)-)).)))-))--)...(((((....)))))......)))) ( -20.70, z-score = -1.33, R) >droMoj3.scaffold_6540 5524642 93 - 34148556 --ACUUGCUGUAGUUAACUGUUGUUGGCUUCACUAAACUCCCGUCGUUUAUAUAUUGUAAUUUUAAGCUCAAUGACGGGUGCAAAAUCGAACAAC------- --..((((((.(((((((....))))))).)).......(((((((((......(((......)))....))))))))).))))...........------- ( -21.90, z-score = -2.10, R) >droVir3.scaffold_13047 8730737 92 - 19223366 --ACUUGCUGUACUCAACUGUUGAUGCCUCUACUGAGCAGGCAUAAUAUAUUUAUAGCAAUUCCGAAUUCAAUGUCGGGUGCAAAACCAAGCAG-------- --..((((((((......(((((.(((((((....)).))))))))))....)))))))).(((((........)))))(((........))).-------- ( -22.50, z-score = -1.41, R) >droGri2.scaffold_14624 4018641 100 - 4233967 --ACUUGCUGCACAUAACUGUUGAUGCCAUGAGAGCACACAAAUAAUAUAUUUAUAGUAAUUCCAAAUGUAAAGAUGAGUGCAAAACAACUGAACAAAGAAC --.((((..(((((.......)).)))..)))).((((.((.....(((((((..(....)...)))))))....)).)))).................... ( -11.70, z-score = 0.97, R) >consensus __AACUGAUGACCCAGUCCGGCAACUCCC_CCCCUUUUAUACCCACUUCGAUUGCCUCAAACUCGAACU_______GAGUGCAAAAUUGCACAA_AUCCAAA ..............................................................................((((......)))).......... ( -3.48 = -3.91 + 0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:30:03 2011