| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,311,188 – 16,311,285 |
| Length | 97 |
| Max. P | 0.958057 |

| Location | 16,311,188 – 16,311,285 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Shannon entropy | 0.33326 |
| G+C content | 0.41044 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.94 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
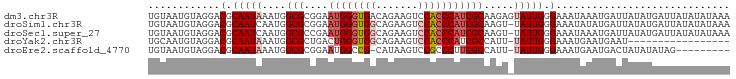
>dm3.chr3R 16311188 97 + 27905053 UGUAAUGUAGGACGCAAUAAAUGGCGCGGAAUGGGUGACAGAAGUCCACCCAUCGCAAGAGUAUUGGGAAAUAAAUGAUUAUAUGAUUAUAUAUAAA ............(.(((((....(((....(((((((((....)).)))))))))).....))))).)..(((.((((((....)))))).)))... ( -21.70, z-score = -1.51, R) >droSim1.chr3R 22353685 96 - 27517382 UGUAAUGUAGGACGCAAUCAAUGGCGCGGAAUGGGUGGCAGAAGUCCACCCAUCGCAAGU-UAUUGGGAAAUAUAUGAUUAUAUGAUUAUAUAUAAA .................(((((((((((..(((((((((....).)))))))))))..))-))))))...((((((((((....))))))))))... ( -28.80, z-score = -2.80, R) >droSec1.super_27 372984 96 - 799419 UGUAAUGUAGGACGCAAUCAAUGGCGCCGAAUGGGUGGCAGAAGUCCACCCAUCGCAAGU-UAUUGGGAAAUAAAUGAUUAUAUGAUUAUAUAUAAA .................((((((((..(((.((((((((....).))))))))))...))-))))))...(((.((((((....)))))).)))... ( -24.80, z-score = -1.51, R) >droYak2.chr3R 5089811 79 + 28832112 UGCAAUGUAGGACGCAAUAAAUGGCGCUGACUGGGUGGCAGAAGUCCACCCAUCGCCAUU-UAUUGGGAAAUGAAUGAAU----------------- ............(.((((((((((((.....((((((((....).))))))).)))))))-))))).)............----------------- ( -29.90, z-score = -3.63, R) >droEre2.scaffold_4770 12385743 86 - 17746568 UGUAAUGUAGGACGCAAUAAAUGGCGCGGAAUGGCCG-CAUAAGUCCGCCCUUCGCCAUU-UAUUGGGAAAUGAAUGACUAUAUAUAG--------- ....((((((..(.((((((((((((.((..(((.(.-.....).)))..)).)))))))-))))).)..........))))))....--------- ( -24.50, z-score = -1.46, R) >consensus UGUAAUGUAGGACGCAAUAAAUGGCGCGGAAUGGGUGGCAGAAGUCCACCCAUCGCAAGU_UAUUGGGAAAUAAAUGAUUAUAUGAUUAUAUAUAAA ............(.(((((....(((....((((((((.......))))))))))).....))))).)............................. (-14.86 = -15.94 + 1.08)

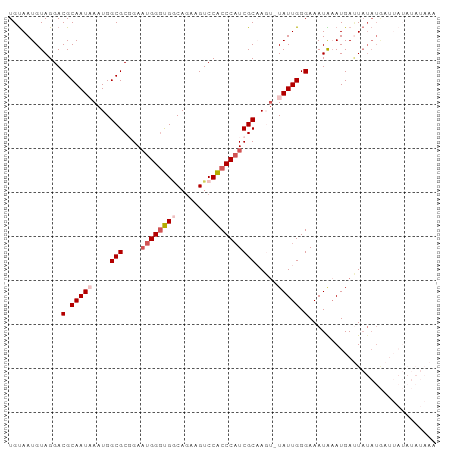
| Location | 16,311,188 – 16,311,285 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Shannon entropy | 0.33326 |
| G+C content | 0.41044 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
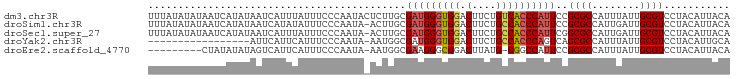
>dm3.chr3R 16311188 97 - 27905053 UUUAUAUAUAAUCAUAUAAUCAUUUAUUUCCCAAUACUCUUGCGAUGGGUGGACUUCUGUCACCCAUUCCGCGCCAUUUAUUGCGUCCUACAUUACA .((((((......))))))............(((((....(((((((((((.((....)))))))))..)))).....))))).............. ( -17.90, z-score = -2.11, R) >droSim1.chr3R 22353685 96 + 27517382 UUUAUAUAUAAUCAUAUAAUCAUAUAUUUCCCAAUA-ACUUGCGAUGGGUGGACUUCUGCCACCCAUUCCGCGCCAUUGAUUGCGUCCUACAUUACA ........((((((((((....))))..........-....(((((((((((.(....)))))))))..))).....)))))).............. ( -19.90, z-score = -1.86, R) >droSec1.super_27 372984 96 + 799419 UUUAUAUAUAAUCAUAUAAUCAUUUAUUUCCCAAUA-ACUUGCGAUGGGUGGACUUCUGCCACCCAUUCGGCGCCAUUGAUUGCGUCCUACAUUACA .((((((......)))))).................-......(((((((((.(....)))))))))).(((((........))))).......... ( -21.60, z-score = -2.09, R) >droYak2.chr3R 5089811 79 - 28832112 -----------------AUUCAUUCAUUUCCCAAUA-AAUGGCGAUGGGUGGACUUCUGCCACCCAGUCAGCGCCAUUUAUUGCGUCCUACAUUGCA -----------------..............(((((-(((((((.(((((((.(....)))))))).....)))))))))))).............. ( -30.00, z-score = -4.91, R) >droEre2.scaffold_4770 12385743 86 + 17746568 ---------CUAUAUAUAGUCAUUCAUUUCCCAAUA-AAUGGCGAAGGGCGGACUUAUG-CGGCCAUUCCGCGCCAUUUAUUGCGUCCUACAUUACA ---------......................(((((-(((((((...(((.(.(....)-).)))......)))))))))))).............. ( -22.60, z-score = -1.68, R) >consensus UUUAUAUAUAAUCAUAUAAUCAUUUAUUUCCCAAUA_ACUUGCGAUGGGUGGACUUCUGCCACCCAUUCCGCGCCAUUUAUUGCGUCCUACAUUACA ...........................................(((((((((.(....))))))))))..((((........))))........... (-13.62 = -13.98 + 0.36)
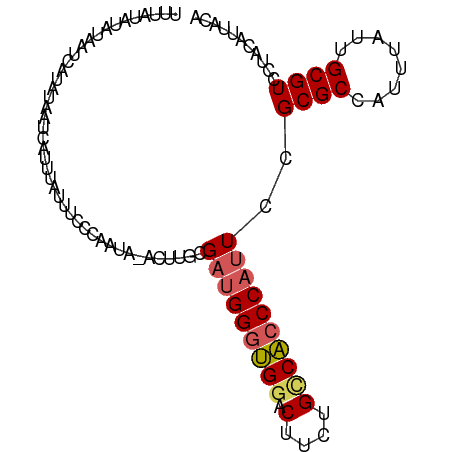
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:59 2011