| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,285,355 – 16,285,456 |
| Length | 101 |
| Max. P | 0.919759 |

| Location | 16,285,355 – 16,285,456 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.09 |
| Shannon entropy | 0.27707 |
| G+C content | 0.48593 |
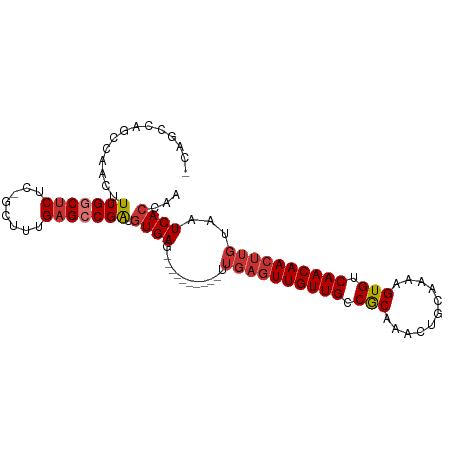
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -20.97 |
| Energy contribution | -22.00 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
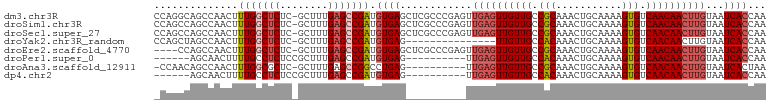
>dm3.chr3R 16285355 101 + 27905053 CCAGGCAGCCAACUUUGGCUCUC-GCUUUGAGCCGAUGUGAGCUCGCCCGAGUUGAGUUGUUGCCGCAAACUGCAAAAGUGUCAACAACUUGUAAUCACCAA ...((((((..((.(((((((..-.....))))))).))..))).))).((..((((((((((.(((...........))).))))))))))...))..... ( -36.10, z-score = -2.84, R) >droSim1.chr3R 22325284 101 - 27517382 CCAGCCAGCCAACUUUGGCUCUC-GCUUUGAGCCGAUGUGAGCUCGCCCGAGUUGAGUUGUUGCCGCAAACUGCAAAAGUGUCAACAACUUGUAAUCACCAA ...((.(((..((.(((((((..-.....))))))).))..))).))..((..((((((((((.(((...........))).))))))))))...))..... ( -33.40, z-score = -2.63, R) >droSec1.super_27 346408 101 - 799419 CCAGCCAGCCAACUUUGGCUCUC-GCUUUGAGCCGAUGUGAGCUCGCCCGAGUUGAGUUGUUGCCGCAAACUGCAAAAGUGUCAACAACUUGUAAUCACCAA ...((.(((..((.(((((((..-.....))))))).))..))).))..((..((((((((((.(((...........))).))))))))))...))..... ( -33.40, z-score = -2.63, R) >droYak2.chr3R_random 222473 86 + 3277457 CCAGCUAGCCAACUUUGGCUCUC-GCUUUGAGCCGAUGUGAG---------------UUGUUGCCACAAACUGCAAAAGUGUCAACAACUUGUAAUCACCAA ..............(((((((..-.....)))))))((..((---------------((((((.(((...........))).))))))))..))........ ( -25.70, z-score = -2.37, R) >droEre2.scaffold_4770 12359055 97 - 17746568 ----CCAGCCAACUUUGGCUCUC-GCUUUGAGCCGAUGUGAGCUCGCCCGAGUUGAGUUGUUGCCGCAAACUGCAAAAGUGUCAACAACUUGUAAUCACCAA ----..(((..((.(((((((..-.....))))))).))..))).....((..((((((((((.(((...........))).))))))))))...))..... ( -30.00, z-score = -2.10, R) >droPer1.super_0 6858426 86 - 11822988 ------AGCAACUUUUGCCUCUCCGCUUUGAGCCGAUGUGAG----------UUGAGUUGUUGCCACAAACUGCAAAAGUGUCAACAACUUGUAAUCACCAA ------........(((.(((........))).))).((((.----------.((((((((((.(((...........))).))))))))))...))))... ( -21.40, z-score = -1.28, R) >droAna3.scaffold_12911 2598928 90 - 5364042 -CCAACAGCCAACUUUGGCGCUC-GCUUUGAGCCGGCCUGAG----------UUGAGUUGUUGCCGCAAACUGCAAAAGUGUCAACAACUUGUAAUCACUAA -.(((((((((((((.(((((((-.....))))..))).)))----------))).)))))))..((.....))...((((...((.....))...)))).. ( -30.30, z-score = -2.45, R) >dp4.chr2 15448744 86 + 30794189 ------AGCAACUUUUGCCUCUCCGCUUUGAGCCGAUGUGAG----------UUGAGUUGUUGCCACAAACUGCAAAAGUGUCAACAACUUGUAAUCACCAA ------........(((.(((........))).))).((((.----------.((((((((((.(((...........))).))))))))))...))))... ( -21.40, z-score = -1.28, R) >consensus _CAGCCAGCCAACUUUGGCUCUC_GCUUUGAGCCGAUGUGAG__________UUGAGUUGUUGCCGCAAACUGCAAAAGUGUCAACAACUUGUAAUCACCAA ..............(((((((........))))))).((((((((....)))).(((((((((.(((...........))).)))))))))....))))... (-20.97 = -22.00 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:56 2011