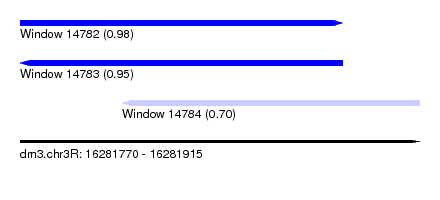
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,281,770 – 16,281,915 |
| Length | 145 |
| Max. P | 0.976011 |

| Location | 16,281,770 – 16,281,887 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Shannon entropy | 0.12619 |
| G+C content | 0.39935 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -27.02 |
| Energy contribution | -26.56 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
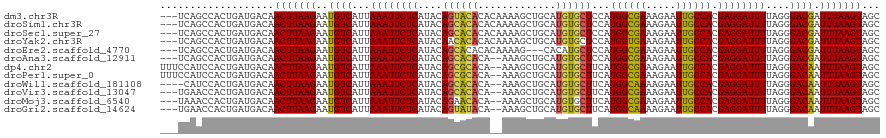
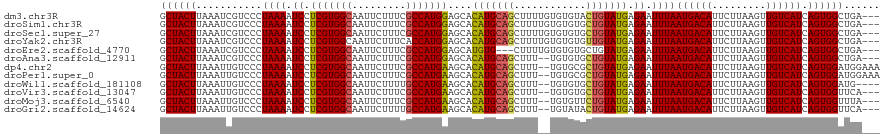
>dm3.chr3R 16281770 117 + 27905053 ---UCAGCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGUACACACAAAAGCUGCAUGUGCUCCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACGAUUUAAGUAGC ---((((...)))).....(((((((..((((...((((((((....((((((..((.....))..))))))...((((((.....)))))).))))))))....)))).)))))))... ( -29.30, z-score = -2.00, R) >droSim1.chr3R 22301785 117 - 27517382 ---UCAGCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCACACACAAAAGCUGCAUGUGCUCCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACGAUUUAAGUAGC ---((((...)))).....(((((((..((((...((((((((....((((((..((.....))..))))))...((((((.....)))))).))))))))....)))).)))))))... ( -31.30, z-score = -2.55, R) >droSec1.super_27 342874 117 - 799419 ---UCAGCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCACACACAAAAGCUGCAUGUGCUCCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACGAUUUAAGUAGC ---((((...)))).....(((((((..((((...((((((((....((((((..((.....))..))))))...((((((.....)))))).))))))))....)))).)))))))... ( -31.30, z-score = -2.55, R) >droYak2.chr3R 5055156 117 + 28832112 ---UCAGCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAACACACACAAAAGCUGCAUGUGCUCCAUGGUGAAAGAAUUGCCACGAGGAUUUUAGGGACGAUUUAAGUAGC ---((((...)))).....(((((((..((((...((((((((......((((..((.....))..)))).....(((..(.....)..))).))))))))....)))).)))))))... ( -24.60, z-score = -0.65, R) >droEre2.scaffold_4770 12355413 114 - 17746568 ---UCAGCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCACACACAAAAG---CACAUGCUCCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACGAUUUAAGUAGC ---((((...)))).....(((((((..((((...((((((((................((---(....)))...((((((.....)))))).))))))))....)))).)))))))... ( -25.70, z-score = -2.01, R) >droAna3.scaffold_12911 2595814 115 - 5364042 ---UCAGCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCACACA--AAAGCUGCAUGUGCUCCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACGAUUUAAGUAGC ---((((...)))).....(((((((..((((...((((((((....((((((((--.....))..))))))...((((((.....)))))).))))))))....)))).)))))))... ( -29.60, z-score = -2.05, R) >dp4.chr2 15445986 118 + 30794189 UUUCCAUCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCGCACA--AAAGCUGCAUGUGCUUCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACAAUUUAAGUAGC ....((((....))))...(((((((..((((...((((((((.((((((((...--...)).)).)))).....((((((.....)))))).))))))))....)))).)))))))... ( -31.40, z-score = -2.51, R) >droPer1.super_0 6855627 118 - 11822988 UUUCCAUCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCGCACA--AAAGCUGCAUGUGCUUCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACAAUUUAAGUAGC ....((((....))))...(((((((..((((...((((((((.((((((((...--...)).)).)))).....((((((.....)))))).))))))))....)))).)))))))... ( -31.40, z-score = -2.51, R) >droWil1.scaffold_181108 2080967 114 - 4707319 ----CAUCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCACACA--AAAGCUGCAUGUGCUUCAUGGCAAAAGAAUUGCCACGAGGAUUUUAGGGACAAUUUAAGUAGC ----((((....))))...(((((((..((((...((((((((....((((((((--.....))..))))))...((((((.....)))))).))))))))....)))).)))))))... ( -31.50, z-score = -3.32, R) >droVir3.scaffold_13047 8674927 115 - 19223366 ---UGAACCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCACACA--AAAGCUGCAUGUGCUUCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACAAUUUAAGUAGC ---................(((((((..((((...((((((((....((((((((--.....))..))))))...((((((.....)))))).))))))))....)))).)))))))... ( -29.30, z-score = -2.53, R) >droMoj3.scaffold_6540 5473519 115 - 34148556 ---UAAACCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGAACACA--AAAGCUGCAUGUGCUUCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACAAUUUAAGUAGC ---................(((((((..((((...((((((((.....(((((((--.........)))).))).((((((.....)))))).))))))))....)))).)))))))... ( -27.70, z-score = -2.63, R) >droGri2.scaffold_14624 3962043 115 - 4233967 ---UGAACCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGUAUACA--AAAGCUGCAUGUGCUUCAUGGCAAAAGAAUUGCCACGAGGAUUUUAGGGACAAUUUAAGUAGC ---................(((((((..((((...((((((((...((((.....--...))))...........((((((.....)))))).))))))))....)))).)))))))... ( -26.60, z-score = -1.84, R) >consensus ___UCAGCCACUGAUGACAACUUAAGAAUGUCAUUAAAUUCUCAUACAGCACACA__AAAGCUGCAUGUGCUCCAUGGCGAAAGAAUUGCCACGAGGAUUUUAGGGACAAUUUAAGUAGC ...................(((((((..((((...((((((((....((((((.............))))))...((((((.....)))))).))))))))....)))).)))))))... (-27.02 = -26.56 + -0.46)
| Location | 16,281,770 – 16,281,887 |
|---|---|
| Length | 117 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.03 |
| Shannon entropy | 0.12619 |
| G+C content | 0.39935 |
| Mean single sequence MFE | -32.61 |
| Consensus MFE | -25.81 |
| Energy contribution | -25.80 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
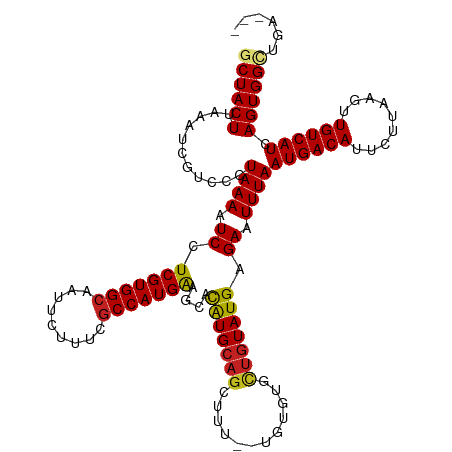
>dm3.chr3R 16281770 117 - 27905053 GCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUUUGUGUGUACUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGCUGA--- .........................(((((((.........)))))))(((((((((((...))))))))((((.((((.(((((((.(.....)))))))).)))))))).)))..--- ( -28.90, z-score = -1.24, R) >droSim1.chr3R 22301785 117 + 27517382 GCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUUUGUGUGUGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGCUGA--- ((((((...................(((((((.........)))))))(((((((((((...)))))))))))(.((((.(((((((.(.....)))))))).)))))))))))...--- ( -35.20, z-score = -2.89, R) >droSec1.super_27 342874 117 + 799419 GCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUUUGUGUGUGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGCUGA--- ((((((...................(((((((.........)))))))(((((((((((...)))))))))))(.((((.(((((((.(.....)))))))).)))))))))))...--- ( -35.20, z-score = -2.89, R) >droYak2.chr3R 5055156 117 - 28832112 GCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCACCAUGGAGCACAUGCAGCUUUUGUGUGUGUUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGCUGA--- ((((((.............((((.(((((((........)))).(((.(((((((((((...))))))))))).)))))).)))).((((((.........)))))).))))))...--- ( -29.80, z-score = -1.51, R) >droEre2.scaffold_4770 12355413 114 + 17746568 GCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCAUGUG---CUUUUGUGUGUGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGCUGA--- ((((((...................(((((((.........)))))))((((((..---(....)..))))))(.((((.(((((((.(.....)))))))).)))))))))))...--- ( -31.80, z-score = -2.59, R) >droAna3.scaffold_12911 2595814 115 + 5364042 GCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUU--UGUGUGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGCUGA--- ((((((((((((...........(..((((((.........))))))..)..((((((((...--.....)))))))).).)))))((((((.........)))))).))))))...--- ( -33.40, z-score = -2.62, R) >dp4.chr2 15445986 118 - 30794189 GCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGAAGCACAUGCAGCUUU--UGUGCGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGAUGGAAA ((.((((((..((((..((((.((.(((((((.........)))))))....((((((((...--.....)))))))).)).))))..))))...)))))).))((((....)))).... ( -34.40, z-score = -2.87, R) >droPer1.super_0 6855627 118 + 11822988 GCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGAAGCACAUGCAGCUUU--UGUGCGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGAUGGAAA ((.((((((..((((..((((.((.(((((((.........)))))))....((((((((...--.....)))))))).)).))))..))))...)))))).))((((....)))).... ( -34.40, z-score = -2.87, R) >droWil1.scaffold_181108 2080967 114 + 4707319 GCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUUGCCAUGAAGCACAUGCAGCUUU--UGUGUGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGAUG---- ((.((((((..((((..((((.((.(((((((((.....)))))))))....((((((((...--.....)))))))).)).))))..))))...)))))).))((((....))))---- ( -37.10, z-score = -4.53, R) >droVir3.scaffold_13047 8674927 115 + 19223366 GCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGAAGCACAUGCAGCUUU--UGUGUGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGUUCA--- ((((((((((((.............(((((((.........)))))))....((((((((...--.....))))))))..))))))((((((.........)))))).))))))...--- ( -32.70, z-score = -3.25, R) >droMoj3.scaffold_6540 5473519 115 + 34148556 GCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGAAGCACAUGCAGCUUU--UGUGUUCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGUUUA--- ((((((((((((.((..........(((((((.........)))))))((((((.........--))))))......)).))))))((((((.........)))))).))))))...--- ( -27.50, z-score = -2.15, R) >droGri2.scaffold_14624 3962043 115 + 4233967 GCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUUGCCAUGAAGCACAUGCAGCUUU--UGUAUACUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGUUCA--- ((((((((((((.............(((((((((.....)))))))))....(((((((....--......)))))))..))))))((((((.........)))))).))))))...--- ( -30.90, z-score = -3.20, R) >consensus GCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGAAGCACAUGCAGCUUU__UGUGUGCUGUAUGAGAAUUUAAUGACAUUCUUAAGUUGUCAUCAGUGGCUGA___ ((((((...........((((.((.(((((((.........)))))))....(((((((............))))))).)).))))((((((.........)))))).))))))...... (-25.81 = -25.80 + -0.01)
| Location | 16,281,807 – 16,281,915 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.59 |
| Shannon entropy | 0.27688 |
| G+C content | 0.46120 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.16 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
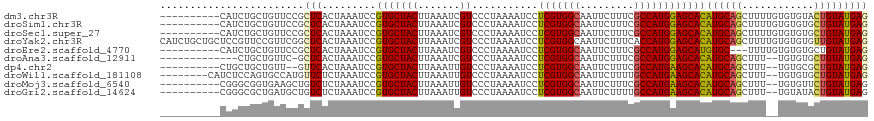
>dm3.chr3R 16281807 108 - 27905053 ----------CAUCUGCUGUUCCGCUCACUAAAUCCGUGCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUUUGUGUGUACUGUAUGAG ----------.......((((((...(((.......)))..........................(((((.........)))))))))))(((((((..(......)..))))))).. ( -21.30, z-score = -0.48, R) >droSim1.chr3R 22301822 108 + 27517382 ----------CAUCUGCUGUUCCGCUCACUAAAUCCGUGCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUUUGUGUGUGCUGUAUGAG ----------.....((......))..........(((((.......................(((((((.........)))))))(((((((((((...)))))))))))))))).. ( -26.10, z-score = -1.78, R) >droSec1.super_27 342911 108 + 799419 ----------CAUCUGCUGUUCCGCUCACUAAAUCCGUGCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUUUGUGUGUGCUGUAUGAG ----------.....((......))..........(((((.......................(((((((.........)))))))(((((((((((...)))))))))))))))).. ( -26.10, z-score = -1.78, R) >droYak2.chr3R 5055193 118 - 28832112 CAUCUGCUGCUCCGUUCCGUUCGGCUCACUAAAUCCGUGCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCACCAUGGAGCACAUGCAGCUUUUGUGUGUGUUGUAUGAG .......((((((((......(((..........)))((((((......................))))))...........))))))))((((((((..........)))))))).. ( -26.25, z-score = -1.09, R) >droEre2.scaffold_4770 12355450 105 + 17746568 ----------CAUCUGCUGUUCCGCUCACUAAAUCCGUGCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCAUGUGC---UUUUGUGUGUGCUGUAUGAG ----------.....((......))..........(((((.......................(((((((.........)))))))((((((..(---....)..))))))))))).. ( -22.70, z-score = -1.52, R) >droAna3.scaffold_12911 2595851 102 + 5364042 -------------CUGCUGUUC-GCUCACUAAAUCCGUGCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUU--UGUGUGCUGUAUGAG -------------.........-((((((.......))..........................((((((.........)))))))))).((((((((...--.....)))))))).. ( -25.60, z-score = -1.99, R) >dp4.chr2 15446026 104 - 30794189 ----------CUGCUGCUGUU--GUUCACUAAAUCCGUGCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGAAGCACAUGCAGCUUU--UGUGCGCUGUAUGAG ----------....((((...--((.(((.......)))..))....................(((((((.........)))))))))))((((((((...--.....)))))))).. ( -25.90, z-score = -2.05, R) >droWil1.scaffold_181108 2081003 108 + 4707319 --------CAUCUCCAGUGCCAUGUUCUCUAAAUCCGUGCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUUGCCAUGAAGCACAUGCAGCUUU--UGUGUGCUGUAUGAG --------..................(((.......(((((((.......))...........(((((((((.....))))))))))))))(((((((...--.....)))))))))) ( -29.00, z-score = -3.73, R) >droMoj3.scaffold_6540 5473556 106 + 34148556 ----------CGGGCGGUGAAGCUGUCUCUAAAUCCGUGCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGAAGCACAUGCAGCUUU--UGUGUUCUGUAUGAG ----------.(((((..((((((((..........(((((((.......))...........(((((((.........))))))))))))..))))))))--..)))))........ ( -26.50, z-score = -1.30, R) >droGri2.scaffold_14624 3962080 106 + 4233967 ----------CGGGCGCUGAUGCUGUCUCUAAAUCCGUGCUACUUAAAUUGUCCCUAAAAUCCUCGUGGCAAUUCUUUUGCCAUGAAGCACAUGCAGCUUU--UGUAUACUGUAUGAG ----------..(((((.(((...........))).)))))......................(((((((((.....)))))))))....(((((((....--......))))))).. ( -27.90, z-score = -2.23, R) >consensus __________CAUCUGCUGUUCCGCUCACUAAAUCCGUGCUACUUAAAUCGUCCCUAAAAUCCUCGUGGCAAUUCUUUCGCCAUGGAGCACAUGCAGCUUU__UGUGUGCUGUAUGAG ........................(((.........(((((((.......))...........(((((((.........))))))))))))((((((............))))))))) (-18.28 = -18.16 + -0.12)
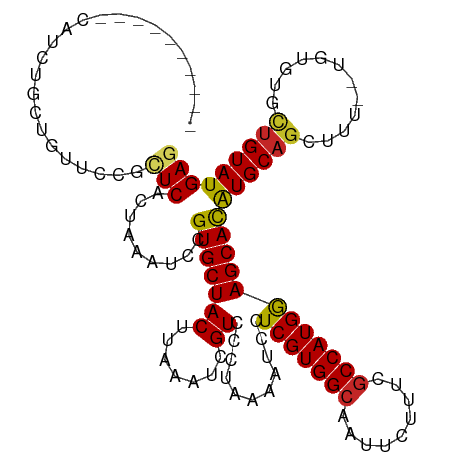
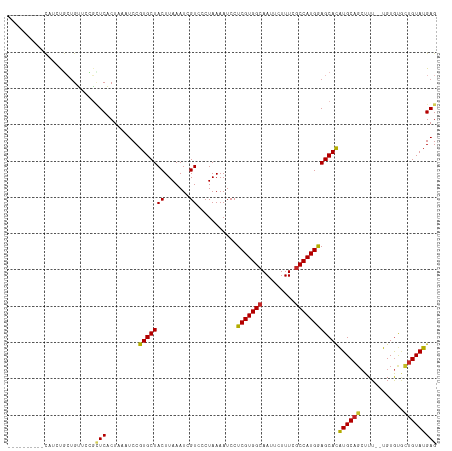
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:55 2011