| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,268,843 – 16,268,971 |
| Length | 128 |
| Max. P | 0.677188 |

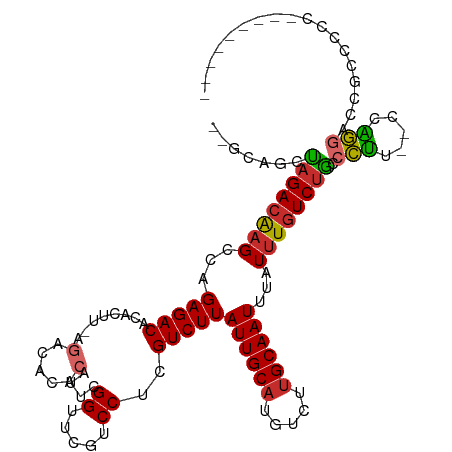
| Location | 16,268,843 – 16,268,937 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.74 |
| Shannon entropy | 0.38709 |
| G+C content | 0.50636 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 16268843 94 + 27905053 --GCAGCUAGACAAGCCAGAGACACAC-----------AUUCGGUUCGUCCUCGUCUUAUUGCAUGUCUUGCAAUUUAUUUGUCUACCCUA--CCAGGACCGCCCCCUU------- --((.((.(((((.((..(((((....-----------...............)))))...)).))))).)).........((((......--...)))).))......------- ( -17.81, z-score = -0.68, R) >droSim1.chr3R 22288696 104 - 27517382 --GCAGCUAGACAAGCCAGAGACACACUU-AGACACACAUUCGGUUCGUCCUCGUCUUAUUGCAUGUCUUGCAAUUUAUUUGUCUGCCCUU--CCAGGACCGCCCCCUU------- --((.((.(((((((...(((((......-.((.......))((.....))..)))))((((((.....))))))...)))))))))((..--...))...))......------- ( -21.20, z-score = -0.85, R) >droSec1.super_27 329765 104 - 799419 --GCAGCUAGACAAGCCAGAGACACACUU-AGACACACAUUCGGUUCGUCCUCGUCUUAUUGCAUGUCUCGCAAUUUAUUUGUCUGCCCUU--CCAGGACCGCCCCCUU------- --((.((.(((((.((..(((((......-.((.......))((.....))..)))))...)).))))).)).........((((......--...)))).))......------- ( -20.50, z-score = -0.68, R) >droYak2.chr3R 5041216 101 + 28832112 --GCAGCUAGACAAGCCAGAGACACACUU-AGACACACAUUCGGUUCGUCCUCGUCUUAUUGCAUGUCUUGCAAUUUAUUUGUCUGCCCUU--CCAGGACCGCCCC---------- --((.((.(((((((...(((((......-.((.......))((.....))..)))))((((((.....))))))...)))))))))((..--...))...))...---------- ( -21.20, z-score = -0.80, R) >droEre2.scaffold_4770 12341865 101 - 17746568 --GCAGCUAGACAAGCCAGAGACACAC-----------AUUCGGUUCGUCCCCGUCUUAUUGCAUGUCUUGCAAUUUAUUUGUCUGCCCUU--CCAGGACCCCCCACCCCCACCCU --...((.(((((((...(((((....-----------...............)))))((((((.....))))))...)))))))))....--...((.....))........... ( -17.51, z-score = -0.84, R) >droAna3.scaffold_12911 2583860 114 - 5364042 GCAGCACCAGACGAGCCAGAGACACACUU-AGACACACAUUUGGCUUGUCCCCGUCUUAUUGCAUGUCUUGCAAUUUAUUUGUCUGCCCUUC-CCGGCCCCGCCCUCAGCCCGCCC ((((((...(((((((((((.........-.........)))))))))))........((((((.....)))))).....)).)))).....-..(((...((.....))..))). ( -29.97, z-score = -2.77, R) >dp4.chr2 15434049 102 + 30794189 GACCAGCUAGACAAGCCAGAGACACACUU-AGACACACAUUUGGCUCGUCCUCGUCUUAUUGCAUGUCUUGCAAUUUAUUUGUCUGCCCUUU-CCAGGAACCCC------------ .....((.((((((((((((.........-.........)))))).............((((((.....))))))....))))))))(((..-..)))......------------ ( -20.57, z-score = -0.85, R) >droPer1.super_0 6843723 104 - 11822988 GACCAGCUAGACAAGCCAGAGACACACUU-AGACACACAUUUGGCUCGUCCUCGUCUUAUUGCAUGUCUUGCAAUUUAUUUGUCUGCCCUU--CCAGGAACCCCCGU--------- .....((.((((((((((((.........-.........)))))).............((((((.....))))))....))))))))....--...((.....))..--------- ( -20.37, z-score = -0.48, R) >droWil1.scaffold_181108 2068925 105 - 4707319 --GCGACCAGACAAGCCAGAGACACACUUUAGACAUACAUUUGGUUUUUCCUCGUCUUAUUGCAUGUCUUGCAAUAUAUAUUUCGGUCUGCCACCCAUACCACCCCC--------- --((((((.((.((((((((...................)))))))).)).......(((((((.....)))))))........)))).))................--------- ( -21.71, z-score = -2.72, R) >consensus __GCAGCUAGACAAGCCAGAGACACACUU_AGACACACAUUCGGUUCGUCCUCGUCUUAUUGCAUGUCUUGCAAUUUAUUUGUCUGCCCUU__CCAGGACCGCCCCC_________ .......((((((((...(((((.(......)..........((.....))..)))))((((((.....))))))...)))))))).(((.....))).................. (-14.14 = -14.18 + 0.04)


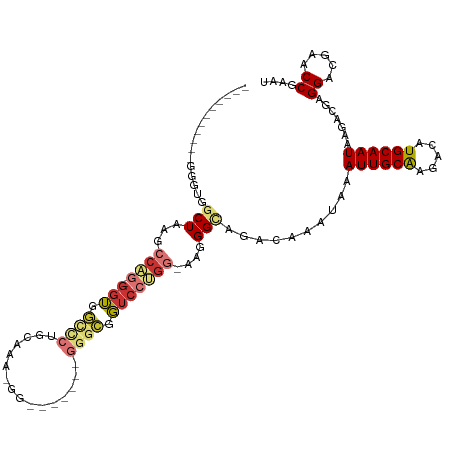
| Location | 16,268,868 – 16,268,971 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.68 |
| Shannon entropy | 0.47786 |
| G+C content | 0.54785 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.46 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.677188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16268868 103 - 27905053 -----GGGGUAGGGUGGCUAAGCCAG-GGUGGCCCUGCAAAAGG-------GGGCGGUCCUGG-UAGGGUAGACAAAUAAAUUGCAAGACAUGCAAUAAGACGAGGACGAACCGAAU -----...(((((((.(((.......-))).))))))).....(-------(..((.((((.(-(.(......)......((((((.....))))))...)).))))))..)).... ( -34.50, z-score = -2.46, R) >droSim1.chr3R 22288731 97 + 27517382 -----------GGGCGGCUAAGCCAG-GGUGGCCCUGCAAAAGG-------GGGCGGUCCUGG-AAGGGCAGACAAAUAAAUUGCAAGACAUGCAAUAAGACGAGGACGAACCGAAU -----------((.(((((...((((-(((.(((((.......)-------)))).)))))))-...)))..........((((((.....))))))..........))..)).... ( -32.90, z-score = -2.28, R) >droSec1.super_27 329800 97 + 799419 -----------GGGCGGCUAAGCCAG-GGUGGCCCUGCAAAAGG-------GGGCGGUCCUGG-AAGGGCAGACAAAUAAAUUGCGAGACAUGCAAUAAGACGAGGACGAACCGAAU -----------((.(((((...((((-(((.(((((.......)-------)))).)))))))-...)))..........((((((.....))))))..........))..)).... ( -31.70, z-score = -1.81, R) >droYak2.chr3R 5041251 94 - 28832112 -----------GGGUGGCUAAGCCAG-GGUGGAUCUGCAA---G-------GGGCGGUCCUGG-AAGGGCAGACAAAUAAAUUGCAAGACAUGCAAUAAGACGAGGACGAACCGAAU -----------.(((.(((...((((-((.....((((..---.-------..))))))))))-...)))..........((((((.....)))))).............))).... ( -25.10, z-score = -1.18, R) >droEre2.scaffold_4770 12341890 104 + 17746568 -----------GCGUGGCUAAGCCAG-GGUGGACCUGCCAAGGGUGGGGGUGGGGGGUCCUGG-AAGGGCAGACAAAUAAAUUGCAAGACAUGCAAUAAGACGGGGACGAACCGAAU -----------((((((((...((((-((....((..((.........))..))...))))))-...))).(.(((.....))))....))))).......((....))........ ( -32.90, z-score = -1.26, R) >droAna3.scaffold_12911 2583897 117 + 5364042 GGCAAAAGGGGGUCUGGCUUGGCAGGAGAUGGUGGGGUGAGGGCGGGCUGAGGGCGGGGCCGGGAAGGGCAGACAAAUAAAUUGCAAGACAUGCAAUAAGACGGGGACAAGCCAAAU (((........(((((((((.((........)).)))).....(.((((........)))).)......)))))......((((((.....)))))).....(....)..))).... ( -30.10, z-score = -1.44, R) >dp4.chr2 15434086 82 - 30794189 ---------------GUCUCGGGCUA-GGGCUU-------------------GGGGUUCCUGGAAAGGGCAGACAAAUAAAUUGCAAGACAUGCAAUAAGACGAGGACGAGCCAAAU ---------------......((((.-(..(((-------------------(..((.(((....)))))..........((((((.....))))))....))))..).)))).... ( -21.70, z-score = -1.26, R) >droPer1.super_0 6843760 89 + 11822988 ---------------GUCUCGGGCUU-GGGGUUUCUGGAC-----------GGGGGUUCCUGG-AAGGGCAGACAAAUAAAUUGCAAGACAUGCAAUAAGACGAGGACGAGCCAAAU ---------------(.((((..(((-(..((((((...(-----------(((....)))).-..)))..)))......((((((.....))))))....))))..)))).).... ( -22.80, z-score = -0.47, R) >consensus ___________GGGUGGCUAAGCCAG_GGUGGCCCUGCAAA_GG_______GGGCGGUCCUGG_AAGGGCAGACAAAUAAAUUGCAAGACAUGCAAUAAGACGAGGACGAACCGAAU ................((...((((....))))...)).............((...(((((.(...(......)......((((((.....))))))....).)))))...)).... (-14.73 = -14.46 + -0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:49 2011