
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,246,427 – 16,246,517 |
| Length | 90 |
| Max. P | 0.893710 |

| Location | 16,246,427 – 16,246,517 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 69.02 |
| Shannon entropy | 0.60186 |
| G+C content | 0.51167 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -14.16 |
| Energy contribution | -15.41 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 16246427 90 - 27905053 CCACUUGCCAUAACAGCAAACUUGGCUACAAAACUGCUGCCAAAGCUCCAGCCGGCAGUGGUGUCCUUGCCAACUAGCAG-AGGAACACAU------ .((((.(((....(((((...(((....)))...))))).....((....)).)))))))((((((((((......))).-))).))))..------ ( -28.30, z-score = -1.39, R) >droAna3.scaffold_12911 2557501 84 + 5364042 CUUUUUGCCAUAACAGCAAACUCUGCCACAAAAG---UGCCAAAGCUACGGUC------GUUGUCCUUGCUGGCUAAGAACAGGGACACUCAG---- ...(((((.......)))))...((.(((....)---)).))..((....)).------..((((((((((.....))..)))))))).....---- ( -16.10, z-score = 1.16, R) >droEre2.scaffold_4770 12319744 90 + 17746568 CCACUUGCCAUAACAGCAAACUUGGCUACAAAACUGCUGCCAAAGCUCCAGCAGGCAGUGGUGUCCUUGCCAACUAACAG-GGGAACACAU------ .((((.(((....(((((...(((....)))...))))).....((....)).)))))))(((((((((........)))-))..))))..------ ( -27.20, z-score = -0.86, R) >droYak2.chr3R_random 2738008 90 - 3277457 CCACUUGCCAUAACAGCAAACUUGGCUACAAAACUGCUGCCAAAGCUCCAGCAGGCAGUGGUGUCCUUGCCAACUUACAG-AGGAACACAU------ .((((.(((....(((((...(((....)))...))))).....((....)).)))))))((((((((...........)-))).))))..------ ( -26.40, z-score = -1.11, R) >droSec1.super_27 307626 74 + 799419 ---------------GUGUG-UUGGCUACAAAACUGCUGCCAAAGCUCCAGCCGGCAGUGGUGUCCUUGCCAACCAGCGG-AGGAACACAU------ ---------------(((((-(((((......((..(((((...((....)).)))))..))......)))..((.....-.)))))))))------ ( -28.50, z-score = -1.56, R) >droSim1.chr3R 22279243 90 + 27517382 CCACUUGCCAUAACAGCAAACUUGGCUACAAAACUGCUGCCAAAGCUCCAGCCGGCAGUGGUGUCCUUGCCAACCAGCGG-AGGAACACAU------ .((((.(((....(((((...(((....)))...))))).....((....)).)))))))((((((((((......)).)-))).))))..------ ( -28.00, z-score = -0.92, R) >droVir3.scaffold_13047 8638134 87 + 19223366 UCAGCCGCCAUAAUACCAAACUAUGCAACACAACUG--GAAACAGUCUGAACU--------UGUCUCUGCCAAACACAGACACACGCACACACAAUU .......................(((....((((((--....)))).))....--------(((((.((.....)).)))))...)))......... ( -15.10, z-score = -2.23, R) >consensus CCACUUGCCAUAACAGCAAACUUGGCUACAAAACUGCUGCCAAAGCUCCAGCCGGCAGUGGUGUCCUUGCCAACUAACAG_AGGAACACAU______ .....................(((((......(((((((((...((....)).)))))))))......)))))........................ (-14.16 = -15.41 + 1.25)

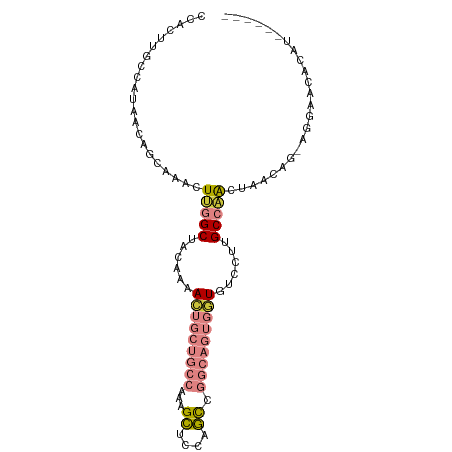

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:45 2011