
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,234,753 – 16,234,870 |
| Length | 117 |
| Max. P | 0.842464 |

| Location | 16,234,753 – 16,234,870 |
|---|---|
| Length | 117 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.74 |
| Shannon entropy | 0.54289 |
| G+C content | 0.51491 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -18.31 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
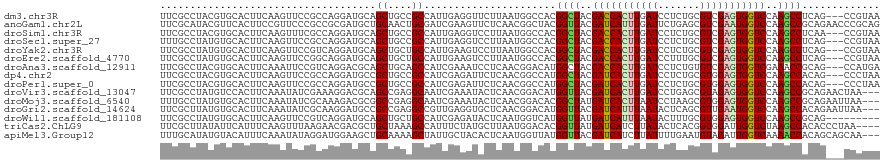
>dm3.chr3R 16234753 117 + 27905053 UUCGCCUACGUGCACUUCAAGUUCCGCCAGGAUGCAGCUGCCGCCAUUGAGGUUCUUAAUGGCCACGGCUACGACCACUUGAUCCUCUGCGUCGAGUGGUCCAAGCCUCAG---CCGUAA ......((((((((........(((....)))))))((((..(((((((((...)))))))))...((((..(((((((((((.......)))))))))))..)))).)))---))))). ( -44.90, z-score = -3.08, R) >anoGam1.chr2L 476691 120 - 48795086 UUCGCAUACGUUCACUUCCGUUCCCGCCGCGAUGCUGCAACUGCGAUCGAAGUUCUCAACGGCUACGGUUACGAUCAUUUGAUUCUGAGCGUCGAAUGGUCCAAGCCGCAGAACCCGCAG ...((....((((....(((((...((..((((((.......)).))))..))....)))))...(((((..(((((((((((.......)))))))))))..)))))..))))..)).. ( -32.90, z-score = 0.08, R) >droSim1.chr3R 22267403 117 - 27517382 UUCGCCUACGUGCACUUCAAGUUUCGCCAGGAUGCAGCUGCCGCCAUUGAGGUCCUUAAUGGCCACGGCUACGACCACUUGAUCCUCUGCGUCGAGUGGUCCAAGCCUCAA---CCGUAA ......((((.((............))...((.(((((((..(((((((((...)))))))))..)))))..(((((((((((.......)))))))))))...)).))..---.)))). ( -39.10, z-score = -1.80, R) >droSec1.super_27 295830 117 - 799419 UUUGCCUAUGUGCACUUCAAGUUCCGCCAGGAUGCAGCUGCCGCCAUUGAGGUCCUUAAUGGCCACGGCUACGACCACUUGAUCCUCUGCGUCGAGUGGUCCAAGCCUCAG---CCGUAA ..(((......)))........(((....)))(((.((((..(((((((((...)))))))))...((((..(((((((((((.......)))))))))))..)))).)))---).))). ( -44.00, z-score = -2.87, R) >droYak2.chr3R 5021215 117 + 28832112 UUCGCCUAUGUGCACUUCAAGUUCCGUCAGGAUGCAGCUGCUGCCAUUGAAGUCCUUAAUGGCCACGGCUACGACCACUUGAUCCUCUGCGUCGAGUGGUCCAAGCCUCAG---CCGUAA ....((((((.(.((.....)).)))).))).(((.((((..((((((((.....))))))))...((((..(((((((((((.......)))))))))))..)))).)))---).))). ( -42.20, z-score = -2.91, R) >droEre2.scaffold_4770 12307552 117 - 17746568 UUCGCCUAUGUGCACUUCAAGUUCCGGCAGGAUGCAGCUGCUGCCAUUGAAGUCCUUAAUGGCCACGGCUACGACCACUUGAUCCUUUGCGUCGAGUGGUCCAAGCCUCAG---CCGUAA ...((......))(((((((....((((((.......))))))...))))))).....(((((...((((..(((((((((((.......)))))))))))..))))...)---)))).. ( -44.60, z-score = -3.06, R) >droAna3.scaffold_12911 2544988 117 - 5364042 UUCGCCUACGUGCACUUCAAAUUCCGUCAGGACGCAGCUGCAGCCAUCGAAAUCCUCAACGGACAUGGCUACGACCACUUGAUCCUCUGUGUCGAGUGGUCGAAACCGCAG---CCAUGA ........((((..........(((....)))....(((((((((((.(...(((.....)))))))))).((((((((((((.......)))))))))))).....))))---))))). ( -42.00, z-score = -3.06, R) >dp4.chr2 15399845 117 + 30794189 UUCGCCUACGUGCACUUCAAGUUCCGCCAGGAUGCCGCUGCCGCCAUCGAGAUUCUCAACGGCCAUGGCUACGAUCACUUGAUCCUCUGCGUGGAGUGGUCCAAGCCACAG---CCCUAA ...((((((((.....(((((((((....))).((((..((((.....(((...)))..))))..)))).......))))))......)))))).(((((....))))).)---)..... ( -34.60, z-score = -0.22, R) >droPer1.super_0 6809309 117 - 11822988 UUCGCCUACGUGCACUUCAAGUUCCGCCAGGAUGCCGCUGCCGCCAUCGAGAUUCUCAACGGCCAUGGCUACGAUCACUUGAUCCUCUGCGUGGAGUGGUCCAAGCCACAG---CCCUAA ...((((((((.....(((((((((....))).((((..((((.....(((...)))..))))..)))).......))))))......)))))).(((((....))))).)---)..... ( -34.60, z-score = -0.22, R) >droVir3.scaffold_13047 8625600 117 - 19223366 UUCGCCUAUGUCCACUUCAAAUAUCGAAAGGACGCAGCCGAGGCAAUCGAAAUACUCAACGGACAUGGUUACGAUCACUUGAUCCUGAGCGUAGAGUGGUCCAAGCCGCAGAACUAA--- ..(((((((((((..........((....)).....((....))................))))))))(((.((((....)))).))))))....(((((....)))))........--- ( -29.70, z-score = -0.56, R) >droMoj3.scaffold_6540 5425716 117 - 34148556 UUUGCCUAUGUGCACUUCAAAUAUCGCAAAGACGCGGCCGAGGCAAUCGAAAUACUCAACGGACACGGCUAUGAUCACUUAAUCCUAAGCGUGGAGUGGUCCAAGCCGCAGAAUUAA--- (((((....(((..(........((((......))))..(((............)))...)..)))((((..((((((((...(......)..))))))))..))))))))).....--- ( -29.20, z-score = -0.18, R) >droGri2.scaffold_14624 3897069 117 - 4233967 UUCGCUUAUGUGCACUUCAAAUAUCGCAAGGAUGCCGCCGAGGCCGUUGAGGUGCUCAACGGACAUGGUUACGAUCAUUUAAUACUCAGCGUUGAAUGGUCCAAGCCACAGAAUUAA--- (((.....((.(((((((((...((....))..(((.....)))..))))))))).)).......(((((..(((((((((((.......)))))))))))..)))))..)))....--- ( -35.60, z-score = -1.69, R) >droWil1.scaffold_181108 2031805 111 - 4707319 UUCGCCUAUGUGCACUUCAAGUUCCGUCAGGAUGCAGCUGCUGCCAUCGAGAUACUCAAUGGUCAUGGUUAUGAUCAUUUAAUACUUUGCGUGGAGUGGUCCAAGCCGCAG--------- ..........((((........(((....))))))).((((.(((((.(((...))).)))))...((((..((((((((..(((.....)))))))))))..))))))))--------- ( -27.20, z-score = 0.91, R) >triCas2.ChLG9 6411056 116 + 15222296 UUCGCUUAUAUUCAUUUCAAGUUUAAGAACGACGCUGCUAAAGCCAUUUCUAUGCUUAAUGGACACGGUUAUGAUCAUCUUAUACUCACGGUGGAUUGGUCUAAGCCACACCCUAA---- ...(((((.(((((((...(((.(((((.....(((.....)))(((..(..((((....)).))..)..)))....))))).)))...))))))).....)))))..........---- ( -20.70, z-score = 0.06, R) >apiMel3.Group12 9057742 117 + 9182753 UUUGCAUAUGUACAUUUCAAAUAUAGGAUGGAAGCUGCAAAAGCUAUUGCUACACUCAAUGGUUAUGGUUACGAUCAUCUUAUUUUGAAUGUAGAUUGGUCAAAACCACAGCAGCAA--- .((((..((.((((((.((((.(((((((((..(..((...((((((((.......))))))))...))..)..))))))))))))))))))).))((((....))))..))))...--- ( -30.50, z-score = -2.25, R) >consensus UUCGCCUAUGUGCACUUCAAGUUCCGCCAGGAUGCAGCUGCCGCCAUUGAGAUCCUCAAUGGCCACGGCUACGAUCACUUGAUCCUCUGCGUCGAGUGGUCCAAGCCACAG___CCGUAA ....................................((....))......................((((..(((((((((((.......)))))))))))..))))............. (-18.31 = -18.03 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:43 2011