
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,208,634 – 16,208,715 |
| Length | 81 |
| Max. P | 0.617973 |

| Location | 16,208,634 – 16,208,715 |
|---|---|
| Length | 81 |
| Sequences | 10 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 79.27 |
| Shannon entropy | 0.43789 |
| G+C content | 0.53630 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -13.30 |
| Energy contribution | -14.26 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
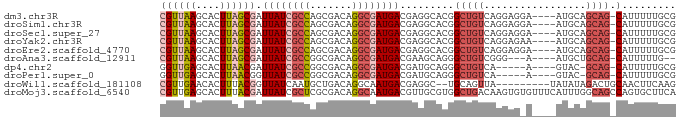
>dm3.chr3R 16208634 81 - 27905053 CGUUAAGCACUUAGCGAUUAUCGCCAGCGACAGGCGAUGACGAGGCACGGCUGUCAGGAGGA----AUGCAGCAG-CAUUUUUGCG ((((((....)))))).((((((((.......))))))))...((((....)))).(.((((----((((....)-))))))).). ( -26.50, z-score = -1.49, R) >droSim1.chr3R 22240710 81 + 27517382 CGUUAAGCACUUAGCGAUUAUCGCCAGCGACAGGCGAUGACGAGGCACGGCUGUCAGGAGGA----AUGCAGCAG-CAUUUUUGCG ((((((....)))))).((((((((.......))))))))...((((....)))).(.((((----((((....)-))))))).). ( -26.50, z-score = -1.49, R) >droSec1.super_27 269433 81 + 799419 CGUUAAGCACUUAGCGAUUAUCGCCAGCGACAGGCGAUGACGAGGCACGGCUGUCAGGAGGA----AUGCAGCAG-CAUUUUUGCG ((((((....)))))).((((((((.......))))))))...((((....)))).(.((((----((((....)-))))))).). ( -26.50, z-score = -1.49, R) >droYak2.chr3R 4986340 81 - 28832112 CGUUAAGCACUUAGCGAUUAUCGCCAGCGACAGGCGAUGACGAGGCACGGCUGUCAGGAGAA----AUGCAGCAG-CAUUUUUGCG ((((((....)))))).((((((((.......))))))))...((((....)))).(.((((----((((....)-))))))).). ( -26.90, z-score = -1.72, R) >droEre2.scaffold_4770 12280463 81 + 17746568 CGUUAAGCACUUAGCGAUUAUCGCCAGCGACAGGCGAUGACGAGGCACGGCUGUCAGGAGGA----AUGCAGCAG-CAUUUUUGCG ((((((....)))))).((((((((.......))))))))...((((....)))).(.((((----((((....)-))))))).). ( -26.50, z-score = -1.49, R) >droAna3.scaffold_12911 2518523 76 + 5364042 CGUUAAGCACUUAGCGAUUAUCGCCGGCGACAGGCGAUGACGAAGCAGGGCUGUCGGG---A----AUGCUGCAG-CAUUUUUG-- ((...(((.(((.((..(((((((((....).))))))))....))))))))..))((---(----((((....)-))))))..-- ( -26.50, z-score = -1.65, R) >dp4.chr2 15373258 75 - 30794189 GGUUGAGCACUUAACGAUUAUCGCCGGCGACAGGCGAUGACGAUGCAGGGCUGUCA-----A----GUAC-GCAG-CAUUUUUGCG .(((((....)))))(.(((((((((....).)))))))))...((((.(((((..-----.----....-))))-)....)))). ( -24.10, z-score = -1.25, R) >droPer1.super_0 6782043 75 + 11822988 GGUUGAGCACUUAACGGUUAUCGCCGGCGACAGGCGAUGACGAUGCAGGGCUGUCA-----A----GUAC-GCAG-CAUUUUUGCG .(((((....))))).((((((((((....).)))))))))...((((.(((((..-----.----....-))))-)....)))). ( -27.60, z-score = -2.18, R) >droWil1.scaffold_181108 2001746 75 + 4707319 CGUUGAACACUUUACGGUUAUCAAUGCUGACAGGCAAUGACGAGGC--UGCAGUUA---------UAUAUAGACUGCAACUUCAAG (((.((....)).)))....(((.((((....)))).))).((((.--(((((((.---------......))))))).))))... ( -20.10, z-score = -2.42, R) >droMoj3.scaffold_6540 5400085 86 + 34148556 CGUUGAGCACUUUACGAUUAUCGCUCGCGACAGGCAAUGACGUUGCGUGGCUGACAAGUGUGUUUCAUUUGGCAGCCAGUGCUUCA ....((((((...............((((((.(.......)))))))((((((.((((((.....)))))).)))))))))))).. ( -30.30, z-score = -1.43, R) >consensus CGUUAAGCACUUAGCGAUUAUCGCCAGCGACAGGCGAUGACGAGGCACGGCUGUCAGGAG_A____AUGCAGCAG_CAUUUUUGCG .....(((......((.((((((((.......)))))))))).......)))...................((((......)))). (-13.30 = -14.26 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:42 2011