| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,191,500 – 16,191,604 |
| Length | 104 |
| Max. P | 0.500000 |

| Location | 16,191,500 – 16,191,604 |
|---|---|
| Length | 104 |
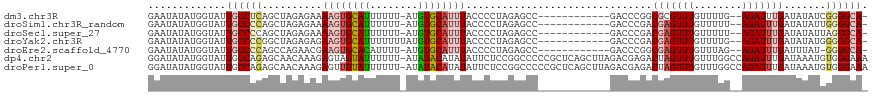
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Shannon entropy | 0.39050 |
| G+C content | 0.41644 |
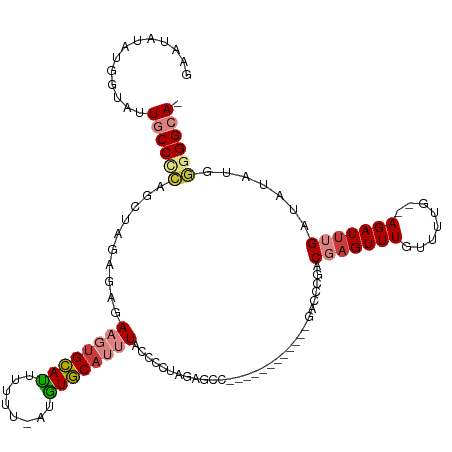
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -14.68 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 16191500 104 + 27905053 GAAUAUAUGGUAUUGCCUCAGCUAGAGAAAAGUGCAUUUUUU-AUGUGCAUUUACCCCUAGAGCC------------GACCCGGCGCGUUUGUUUUG--AGAUUUGAUAUAUCGGGGCA- .............((((((..((((.(..((((((((.....-..))))))))..).)))).(((------------(...))))............--..............))))))- ( -26.30, z-score = -0.70, R) >droSim1.chr3R_random 89380 104 + 1307089 GAAUAUAUGGUAUUGCCCCAGCUAGAGAAAAGUGCAUUUUUU-AUGUGCAUUUACCCCUAGAGCC------------GACCCGACGAGUUUGUUUUU--AGAUUUGAUAUAUUGGGGCA- .............((((((((((((.(..((((((((.....-..))))))))..).))))....------------.......((((((((....)--))))))).....))))))))- ( -29.70, z-score = -3.49, R) >droSec1.super_27 252617 104 - 799419 GAAUAUAUGGUAUUGCCCCAGCUAGAGAAAAGUGCAUUUUUU-AUGUGCAUUUACCCCUAGAGCC------------GACCCGACGAGUUUGUUUUU--AGAUUUGAUAUAUUAGGGCA- .............(((((...((((.(..((((((((.....-..))))))))..).))))....------------.......((((((((....)--)))))))........)))))- ( -24.40, z-score = -2.29, R) >droYak2.chr3R 4967419 105 + 28832112 GAAUAUAUGGUAUUGCCCCGGCUAGAGAGAAGUGCAUUUUUUUAUGUGCAUUUACCCCUAGAGCC------------GACCCGACGAGUUUGUUUUG--AGAUUUGAUAUAUGGGGGCA- .............((((((((((..((..((((((((........))))))))....))..))))------------.......(((((((......--))))))).......))))))- ( -32.40, z-score = -3.23, R) >droEre2.scaffold_4770 12262575 103 - 17746568 GAAUAUAUGGUAUUGCCCCAGCCAGAACGAAGUGCACAUUUU-AUGUGCAUUUACCCCUAGAGCC------------GACCCGGCGAGUUUGUUUAG--AGAUUUGAUUUAU-GGGGCA- .............(((((((..((((.(.((((((((((...-)))))))))).........(((------------(...))))............--.).)))).....)-))))))- ( -29.70, z-score = -2.50, R) >dp4.chr2 15356559 119 + 30794189 GGAUAUAUGGUAUUGCCAGAGCAACAAAGAGUAGUAUUUUUU-AUAUACAUAUAUUCUCCGGCCCCCGCUCAGCUUAGACGAGACUAGUUUGUUUGGCCAGAUUUGAUAAAUGUGGGAAA (((((((((...((((....)))).(((((((...)))))))-.....))))))))).......(((((((((.((.(.(.((((......)))).).).)).)))).....)))))... ( -23.60, z-score = 0.61, R) >droPer1.super_0 6765155 119 - 11822988 GGAUAUAUGGUAUUGCCAGAGCAACAAAGAGUUGUAUUUUUU-AUAUACAUAUAUUCUCCGGCCCCCGCUCAGCUUAGACGAGACUAGUUUGUUUGGCCAGAUUUGAUAAAUGUGGGAAA (((((((((((((.......(((((.....))))).......-)))).))))))))).......(((((((((.((.(.(.((((......)))).).).)).)))).....)))))... ( -25.84, z-score = -0.01, R) >consensus GAAUAUAUGGUAUUGCCCCAGCUAGAGAGAAGUGCAUUUUUU_AUGUGCAUUUACCCCUAGAGCC____________GACCCGACGAGUUUGUUUUG__AGAUUUGAUAUAUGGGGGCA_ .............((((((..........((((((((........))))))))...............................(((((((........))))))).......)))))). (-14.68 = -15.40 + 0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:40 2011