| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,177,183 – 16,177,337 |
| Length | 154 |
| Max. P | 0.809079 |

| Location | 16,177,183 – 16,177,337 |
|---|---|
| Length | 154 |
| Sequences | 9 |
| Columns | 155 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Shannon entropy | 0.34955 |
| G+C content | 0.43869 |
| Mean single sequence MFE | -41.91 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.91 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
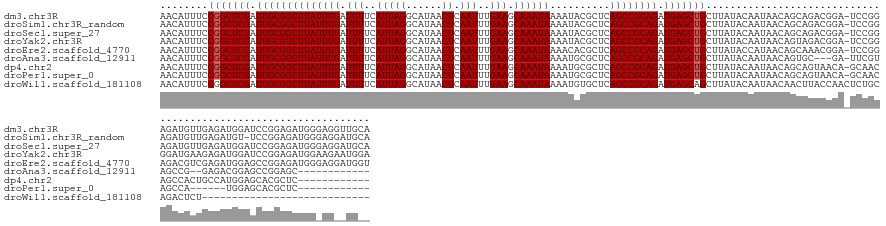
>dm3.chr3R 16177183 154 + 27905053 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUACGCUCAGCCGCAGAUGAGCUGCUUAUACAAUAACAGCAGACGGA-UCCGGAGAUGUUGAGAUGGAUCCGGAGAUGGGAGGUUGCA ..(((((((((.((.(((((((((((((((..........(((.....((.....))(((...........)))..))).))))))))))))...........(((((..((..-..))....))))).))).)).))))))))).......... ( -46.90, z-score = -2.17, R) >droSim1.chr3R_random 74878 153 + 1307089 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUACGCUCAGCCGCAGAUGAGCUGCUUAUACAAUAACAGCAGACGGA-UCCGGAGAUGUUGAGAUGU-UCCGGAGAUGGGAGGAUGCA (((((((((((((((.((((((((((((((.(((..(((((......)).)))..))).))))))..........)))))))).))))(((((...........))))).....-.)))))))))))...((.(-(((.......)))).))... ( -44.60, z-score = -1.69, R) >droSec1.super_27 238283 154 - 799419 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUACGCUCAGCCGCAGAUGAGCUGCUUAUACAAUAACAGCAGACGGA-UCCGGAGAUGUUGAGAUGGAUCCGGAGAUGGGAGGAUGCA ..(((((((((.((.(((((((((((((((..........(((.....((.....))(((...........)))..))).))))))))))))...........(((((..((..-..))....))))).))).)).))))))))).......... ( -46.90, z-score = -2.30, R) >droYak2.chr3R 4953263 154 + 28832112 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUACGCUCAGCCGCAGAUGAGCUGCUUAUACAAUAACAGUAGACGGA-UCCGGGGAUGAAGAGAUGGAUCCGGAGAUGGAAGAAUGGA ..(((((((((((((.((((((((((((((.(((..(((((......)).)))..))).))))))..........)))))))).)))))).....................(((-((((............)))))))))))))).......... ( -45.80, z-score = -2.64, R) >droEre2.scaffold_4770 12247754 154 - 17746568 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAACACGCUCAGCCGCAGAUGAGCUGCUUAUACCAUAACAGCAAACGGA-UCCGGAGACGUCGAGAUGGAGCCGGAGAUGGGAGGAUGGU ..((((((((((((.(((((((((((((((.(((..(((((......)).)))..))).))))))..........)))))).((((.((((.((((...))))))))...((..-..))....))))..))).)))))))))))).......... ( -50.60, z-score = -3.12, R) >droAna3.scaffold_12911 2489274 137 - 5364042 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUGCGCUCAGCCGCAGAUGAGCUGCUUAUACAAUAACAGUGC---GA-UUCGUAGCCG--GAGACGGAGCCGGAGC------------ .....(((((((((....((((((((((((..........(((.....((.....)).(((.......))).....))).))))))))))))...............((---..-......))((--....))))))))))).------------ ( -43.80, z-score = -2.30, R) >dp4.chr2 15341979 142 + 30794189 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUGCGCUCAGCCGCAGAUGAGCUGCUUAUACAAUAACAGCAGUAACA-GCAACAGCCACUGCCAUGGAGCACGCUC------------ ..(((((.(((((.((.((((..(((((((.(((..(((((......)).)))..))).)))))))..)))).))))))).)))))((((((((...........(((((....-((....)).)))))....))))).))).------------ ( -37.86, z-score = -1.11, R) >droPer1.super_0 6750511 136 - 11822988 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUGCGCUCAGCCGCAGAUGAGCUGCUUAUACAAUAACAGCAGUAACA-GCAACAGCCA------UGGAGCACGCUC------------ ..(((((.(((((.((.((((..(((((((.(((..(((((......)).)))..))).)))))))..)))).))))))).))))).((((((...........))))))....-((..((....------))..))......------------ ( -35.00, z-score = -0.94, R) >droWil1.scaffold_181108 1969042 127 - 4707319 AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUGUGCUCAGCCGCAGAUGAGCAGCUUAUACAAUAACAACUUACCAACUCUGCAGACUCU---------------------------- ..........(((((.((((((((....((.....))...((((((.....(((((...)))))....)))))).)))))))).)))))......................................---------------------------- ( -25.70, z-score = -0.50, R) >consensus AACAUUUCCGGCUCGAUUGCGGCUUAUUUGAUUUUCAUUAGGCAUAACUCAAUUUGAAGCAAAUAAAAUACGCUCAGCCGCAGAUGAGCUGCUUAUACAAUAACAGCAGACGGA_UCCGGAGACGUUGAGAUGGAGCCGGAGAUGG_AG__UG__ ........(((((((.((((((((((((((.(((..(((((......)).)))..))).))))))..........)))))))).)))))))................................................................ (-26.80 = -26.91 + 0.11)

| Location | 16,177,183 – 16,177,337 |
|---|---|
| Length | 154 |
| Sequences | 9 |
| Columns | 155 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Shannon entropy | 0.34955 |
| G+C content | 0.43869 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -26.99 |
| Energy contribution | -27.51 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
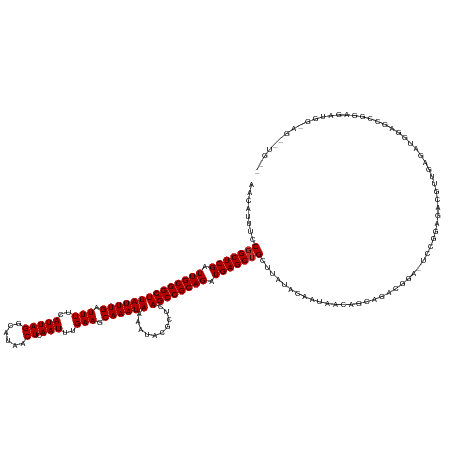
>dm3.chr3R 16177183 154 - 27905053 UGCAACCUCCCAUCUCCGGAUCCAUCUCAACAUCUCCGGA-UCCGUCUGCUGUUAUUGUAUAAGCAGCUCAUCUGCGGCUGAGCGUAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU ..........(((.(((((((((..............)))-)))...((((...........))))((((...((((((((((((.........)))))....((((.((((.....))))...))))...)))))))...)))).))).))).. ( -41.84, z-score = -2.38, R) >droSim1.chr3R_random 74878 153 - 1307089 UGCAUCCUCCCAUCUCCGGA-ACAUCUCAACAUCUCCGGA-UCCGUCUGCUGUUAUUGUAUAAGCAGCUCAUCUGCGGCUGAGCGUAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU .((((.........((((((-.............))))))-((((..((((...........))))((((...((((((((((((.........)))))....((((.((((.....))))...))))...)))))))...)))))))).)))). ( -41.12, z-score = -2.18, R) >droSec1.super_27 238283 154 + 799419 UGCAUCCUCCCAUCUCCGGAUCCAUCUCAACAUCUCCGGA-UCCGUCUGCUGUUAUUGUAUAAGCAGCUCAUCUGCGGCUGAGCGUAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU .((((..(((...((((((((((..............)))-))))...((((((........)))))).....((((((((((((.........)))))....((((.((((.....))))...))))...)))))))...)))..))).)))). ( -42.54, z-score = -2.55, R) >droYak2.chr3R 4953263 154 - 28832112 UCCAUUCUUCCAUCUCCGGAUCCAUCUCUUCAUCCCCGGA-UCCGUCUACUGUUAUUGUAUAAGCAGCUCAUCUGCGGCUGAGCGUAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU .......((((.....(((((((..............)))-)))).....((((........))))((((...((((((((((((.........)))))....((((.((((.....))))...))))...)))))))...)))).))))..... ( -39.74, z-score = -2.63, R) >droEre2.scaffold_4770 12247754 154 + 17746568 ACCAUCCUCCCAUCUCCGGCUCCAUCUCGACGUCUCCGGA-UCCGUUUGCUGUUAUGGUAUAAGCAGCUCAUCUGCGGCUGAGCGUGUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU ..........(((.((((((((.((((.((....)).)))-)......((((((........)))))).....((((((((((((.........)))))....((((.((((.....))))...))))...)))))))...)))))))).))).. ( -45.00, z-score = -2.46, R) >droAna3.scaffold_12911 2489274 137 + 5364042 ------------GCUCCGGCUCCGUCUC--CGGCUACGAA-UC---GCACUGUUAUUGUAUAAGCAGCUCAUCUGCGGCUGAGCGCAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU ------------((....))..(((.((--(((((.(((.-..---(..(((((........)))))..)...((((((((((.(((.......))))))...((((.((((.....))))...))))...))))))).)))))))))).))).. ( -42.30, z-score = -2.16, R) >dp4.chr2 15341979 142 - 30794189 ------------GAGCGUGCUCCAUGGCAGUGGCUGUUGC-UGUUACUGCUGUUAUUGUAUAAGCAGCUCAUCUGCGGCUGAGCGCAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU ------------.(((((..((((((((((((((......-.))))))))))).............((((...((((((((((.(((.......))))))...((((.((((.....))))...))))...)))))))...)))).))).))))) ( -45.60, z-score = -1.38, R) >droPer1.super_0 6750511 136 + 11822988 ------------GAGCGUGCUCCA------UGGCUGUUGC-UGUUACUGCUGUUAUUGUAUAAGCAGCUCAUCUGCGGCUGAGCGCAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU ------------.(((((..(((.------.(((((((((-(..(((..........)))..)))))).....((((((((((.(((.......))))))...((((.((((.....))))...))))...)))))))....))))))).))))) ( -39.90, z-score = -0.88, R) >droWil1.scaffold_181108 1969042 127 + 4707319 ----------------------------AGAGUCUGCAGAGUUGGUAAGUUGUUAUUGUAUAAGCUGCUCAUCUGCGGCUGAGCACAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU ----------------------------....((((...((((.((((((....))).))).))))((((...((((((((((((.........)))))....((((.((((.....))))...))))...)))))))...))))))))...... ( -28.00, z-score = 0.76, R) >consensus __CA__CU_CCAUCUCCGGAUCCAUCUCAACGUCUCCGGA_UCCGUCUGCUGUUAUUGUAUAAGCAGCUCAUCUGCGGCUGAGCGUAUUUUAUUUGCUUCAAAUUGAGUUAUGCCUAAUGAAAAUCAAAUAAGCCGCAAUCGAGCCGGAAAUGUU ....................(((........................((((...........))))((((...((((((((((((.........)))))....((((.((((.....))))...))))...)))))))...)))).)))...... (-26.99 = -27.51 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:38 2011