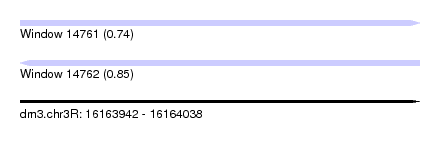
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,163,942 – 16,164,038 |
| Length | 96 |
| Max. P | 0.846857 |

| Location | 16,163,942 – 16,164,038 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.90 |
| Shannon entropy | 0.24658 |
| G+C content | 0.51079 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -21.17 |
| Energy contribution | -22.37 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
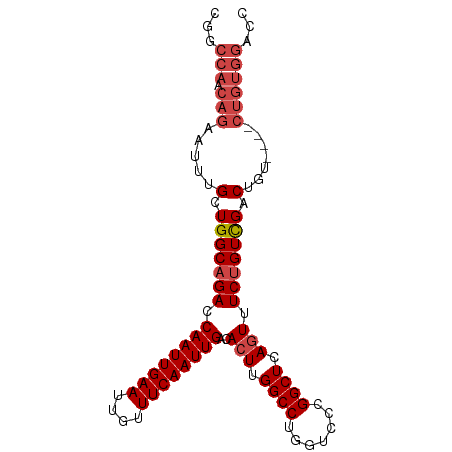
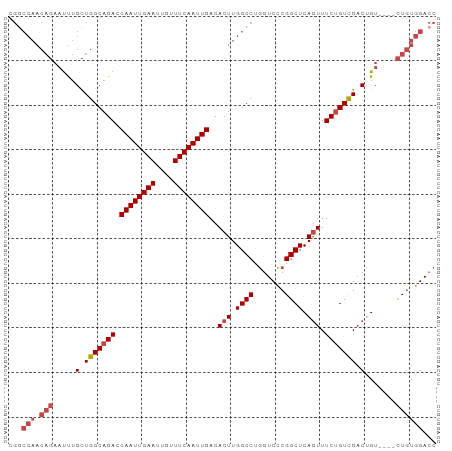
>dm3.chr3R 16163942 96 + 27905053 CGACCAACAGAAUUUGCUGGCAGACCAAUUGAAUUGUUUCAAUUGACACUUGGCCUGGUCCCGGCUCAGUUUCUGUCGACUGUGGAUCUGUGGACC ...(((.((((.((..((((((((.((((((((....))))))))..(((.((((.(....))))).))).)))))))...)..)))))))))... ( -32.90, z-score = -2.39, R) >droSim1.chr3R 22201706 96 - 27517382 CGGCCAACAGAAUUUGCUGGCAGACCAAUUGAAUUGUUUCAAUUGACACUUGGCCUGGUCCCGGCUCAGUUUCUGUCGACUGUGGAUCUGUGGACC .(((((.((((.((..((((((((.((((((((....))))))))..(((.((((.(....))))).))).)))))))...)..))))))))).)) ( -34.50, z-score = -2.41, R) >droSec1.super_27 225041 96 - 799419 CGGCCAACAGAAUUUGCUGGCAGACCAAUUGAAUUGUUUCAAUUGACACUUGGCCUGGUCCCGGCUCAAUUUCUGUCGACUGUGGAUCUGUGGACC .(((((.((((.((..((((((((.((((((((....))))))))....((((((.(....)))).)))..)))))))...)..))))))))).)) ( -32.60, z-score = -2.25, R) >droYak2.chr3R_random 2682636 92 + 3277457 CGGCCAACAGAAUUUGCUGGCAGACCAAUUGAAUUGUUUCAAUUGACACUUGGCCGGGUCCCGGCUCAGUUUCUGUCGACCGU----CUGUGGACC .(((((.((((....(.(((((((.((((((((....))))))))..(((.((((((...)))))).))).))))))).)..)----)))))).)) ( -36.10, z-score = -3.03, R) >droEre2.scaffold_4770 12234862 92 - 17746568 CGGCCAACAGAAUUUGCUGGCAGACCAAUUGAAUUGUUUCAAUUGACACUUGGCCUGGUCCCGGCUCAGUUUCUGUCGACCGU----CUGUGGACC .(((((.((((....(.(((((((.((((((((....))))))))..(((.((((.(....))))).))).))))))).)..)----)))))).)) ( -32.00, z-score = -2.17, R) >droAna3.scaffold_12911 2476226 80 - 5364042 ------------CCGGUUGGCAGAGCAAUUGAAUUGUUUCAAUUGACACUUGGCCCAGUCUCGGCUCAGUUUCCGUUGCCUGU----UUUUGGAGC ------------((((..(((((.(((((((((....)))))))((.(((.((((.......)))).))).))....))))))----).))))... ( -23.00, z-score = -0.75, R) >consensus CGGCCAACAGAAUUUGCUGGCAGACCAAUUGAAUUGUUUCAAUUGACACUUGGCCUGGUCCCGGCUCAGUUUCUGUCGACUGU____CUGUGGACC ...(((.(((.....(.(((((((.((((((((....))))))))..(((.((((.......)))).))).))))))).).......))))))... (-21.17 = -22.37 + 1.19)

| Location | 16,163,942 – 16,164,038 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.90 |
| Shannon entropy | 0.24658 |
| G+C content | 0.51079 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -21.33 |
| Energy contribution | -22.83 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.846857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
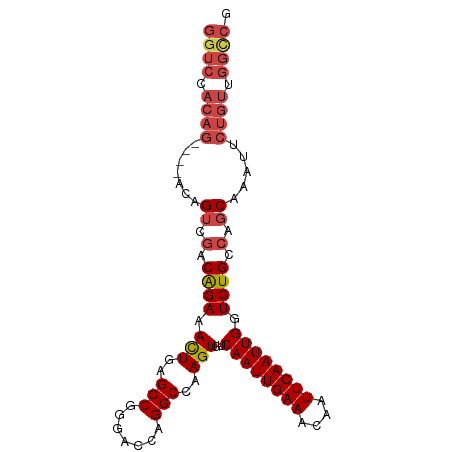
>dm3.chr3R 16163942 96 - 27905053 GGUCCACAGAUCCACAGUCGACAGAAACUGAGCCGGGACCAGGCCAAGUGUCAAUUGAAACAAUUCAAUUGGUCUGCCAGCAAAUUCUGUUGGUCG (..(.(((((....((((........)))).((.((...((((((((.((..(((((...))))))).)))))))))).))....))))).)..). ( -27.70, z-score = -1.08, R) >droSim1.chr3R 22201706 96 + 27517382 GGUCCACAGAUCCACAGUCGACAGAAACUGAGCCGGGACCAGGCCAAGUGUCAAUUGAAACAAUUCAAUUGGUCUGCCAGCAAAUUCUGUUGGCCG ((((.(((((....((((........)))).((.((...((((((((.((..(((((...))))))).)))))))))).))....))))).)))). ( -30.90, z-score = -1.98, R) >droSec1.super_27 225041 96 + 799419 GGUCCACAGAUCCACAGUCGACAGAAAUUGAGCCGGGACCAGGCCAAGUGUCAAUUGAAACAAUUCAAUUGGUCUGCCAGCAAAUUCUGUUGGCCG ((((....))))....((((((((((.(((.((..(((((.(((.....)))(((((((....))))))))))))))...))).)))))))))).. ( -30.40, z-score = -2.00, R) >droYak2.chr3R_random 2682636 92 - 3277457 GGUCCACAG----ACGGUCGACAGAAACUGAGCCGGGACCCGGCCAAGUGUCAAUUGAAACAAUUCAAUUGGUCUGCCAGCAAAUUCUGUUGGCCG .(((....)----))((((((((((((((..(((((...)))))..))).(((((((((....)))))))))............))))))))))). ( -36.90, z-score = -3.54, R) >droEre2.scaffold_4770 12234862 92 + 17746568 GGUCCACAG----ACGGUCGACAGAAACUGAGCCGGGACCAGGCCAAGUGUCAAUUGAAACAAUUCAAUUGGUCUGCCAGCAAAUUCUGUUGGCCG .(((....)----))(((((((((((.(((.((..(((((.(((.....)))(((((((....)))))))))))))))))....))))))))))). ( -34.90, z-score = -3.08, R) >droAna3.scaffold_12911 2476226 80 + 5364042 GCUCCAAAA----ACAGGCAACGGAAACUGAGCCGAGACUGGGCCAAGUGUCAAUUGAAACAAUUCAAUUGCUCUGCCAACCGG------------ .........----...((((..(....).((((.((.(((......))).))(((((((....)))))))))))))))......------------ ( -21.70, z-score = -1.52, R) >consensus GGUCCACAG____ACAGUCGACAGAAACUGAGCCGGGACCAGGCCAAGUGUCAAUUGAAACAAUUCAAUUGGUCUGCCAGCAAAUUCUGUUGGCCG ................((((((((((.(((.((..(((((.(((.....)))(((((((....)))))))))))))))))....)))))))))).. (-21.33 = -22.83 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:36 2011