| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,152,120 – 16,152,170 |
| Length | 50 |
| Max. P | 0.954381 |

| Location | 16,152,120 – 16,152,170 |
|---|---|
| Length | 50 |
| Sequences | 12 |
| Columns | 50 |
| Reading direction | reverse |
| Mean pairwise identity | 75.09 |
| Shannon entropy | 0.59190 |
| G+C content | 0.55500 |
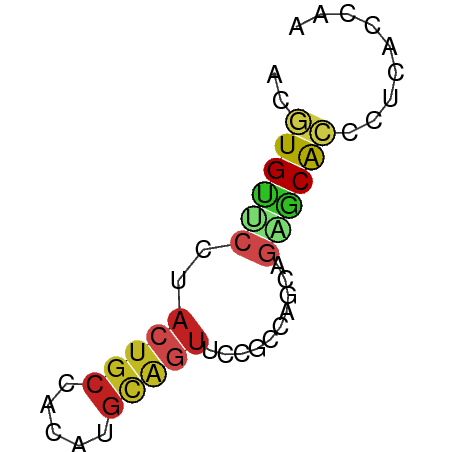
| Mean single sequence MFE | -11.05 |
| Consensus MFE | -6.30 |
| Energy contribution | -6.74 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
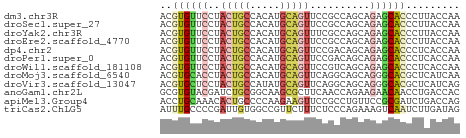
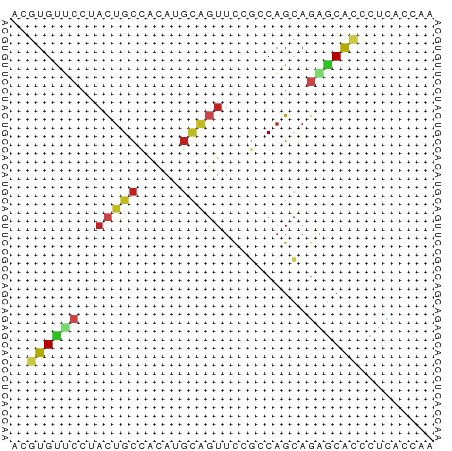
>dm3.chr3R 16152120 50 - 27905053 ACGUGUUCCUACUGCCACAUGCAGUUCCGCCAGCAGAGCACCCUUACCAA ..((((((..(((((.....)))))...(....).))))))......... ( -10.90, z-score = -1.84, R) >droSec1.super_27 213668 50 + 799419 ACGUGUUCCUACUGCCACAUGCAGUUCCGCCAGCAGAGCACCCUUACCAA ..((((((..(((((.....)))))...(....).))))))......... ( -10.90, z-score = -1.84, R) >droYak2.chr3R 4930332 50 - 28832112 ACGUGUUCCUACUGCCACAUGCAGUUUCGCCAGCAGAGCACCCUUACCAA ..((((((..(((((.....)))))...(....).))))))......... ( -10.90, z-score = -1.87, R) >droEre2.scaffold_4770 12223344 50 + 17746568 ACGUGUUCCUACUGCCACAUGCAGUUCCGCCAGCAGAGCACCCUUACCAA ..((((((..(((((.....)))))...(....).))))))......... ( -10.90, z-score = -1.84, R) >dp4.chr2 15316608 50 - 30794189 ACGUGUUCCUACUGCCACAUGCAGUUCCGACAGCAGAGCACCCUCACCAA ..((((((..(((((.....)))))..........))))))......... ( -10.20, z-score = -1.25, R) >droPer1.super_0 6725073 50 + 11822988 ACGUGUUCCUACUGCCACAUGCAGUUCCGACAGCAGAGCACCCUCACCAA ..((((((..(((((.....)))))..........))))))......... ( -10.20, z-score = -1.25, R) >droWil1.scaffold_181108 1942097 50 + 4707319 ACGUGUUCCUACUGCCACAUGCAGUUCCGUCAGCAGAGCACCCUCACCAA ..((((((..(((((.....)))))...(....).))))))......... ( -10.70, z-score = -1.58, R) >droMoj3.scaffold_6540 14561274 50 + 34148556 ACGUGCACCUACUGCCACAUGCAGUUCAGGCAGCAGGGCACGCUCAUCAA .(((((.(((.(((((............))))).))))))))........ ( -20.50, z-score = -3.19, R) >droVir3.scaffold_13047 3168422 50 + 19223366 ACGUGCUCCUACUGCCAUAUGCAGUUCAGGCAGCAGGGCACGCUCAUCAG .(((((.(((.(((((............))))).))))))))........ ( -20.50, z-score = -3.22, R) >anoGam1.chr2L 3088588 50 + 48795086 GCGUGUACGAUCUGCGGCAAGCGCUUCAACCAGAAGAACAACCUGACCAC ((((...((.....))....))))......(((.........)))..... ( -7.10, z-score = 0.97, R) >apiMel3.Group4 6783860 50 - 10796202 ACCUGCAAACACUGCCCCAAGAAGUUCCGCCUGUUCCGCGAUCUGACCAG ....(((.....)))............(((.......))).......... ( -3.40, z-score = 1.20, R) >triCas2.ChLG5 1392453 50 + 18847211 AUUUGCCCCGAUUGUGGCCGUUCUUUCUCCCAGAAAGUCAAUCUUGAUAG .........(((((........(((((.....))))).)))))....... ( -6.40, z-score = -0.21, R) >consensus ACGUGUUCCUACUGCCACAUGCAGUUCCGCCAGCAGAGCACCCUCACCAA ..((((((..(((((.....)))))..........))))))......... ( -6.30 = -6.74 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:33 2011