| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,146,187 – 16,146,295 |
| Length | 108 |
| Max. P | 0.626381 |

| Location | 16,146,187 – 16,146,295 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.19 |
| Shannon entropy | 0.69437 |
| G+C content | 0.66061 |
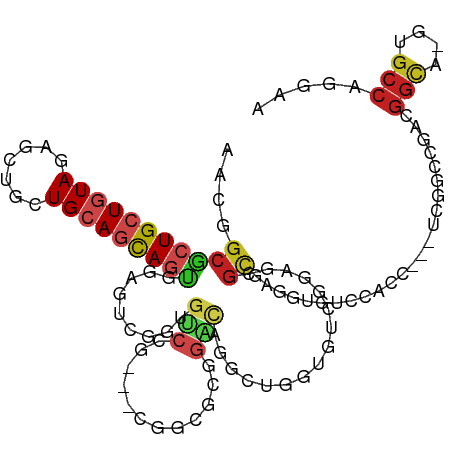
| Mean single sequence MFE | -50.77 |
| Consensus MFE | -15.17 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
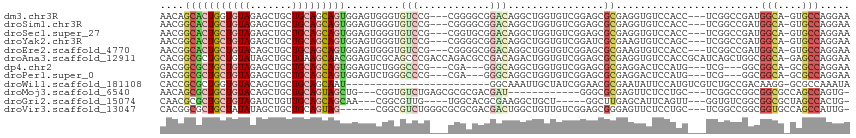
>dm3.chr3R 16146187 108 - 27905053 AACAGCACUGGUGUAGAGCUGCUGCAGCAGUGGAGUGGGUGUCCG---CGGGGCGGACAGGCUGGUGUCGGAGCGCGAGGUGUCCACC---UCGGCCGAUGGCA-GUGCCAGGAA .....(((((.(((((.....))))).)))))...(((.((((((---(...))))))).((((.((((((....((((((....)))---))).)))))).))-)).))).... ( -51.10, z-score = -1.45, R) >droSim1.chr3R 22182695 108 + 27517382 AACGGCACUGCUGUAGAGCUGCUGCAGCAGUGGAGUGGGUGUCCG---CGGGGCGGACAGGCUGGUGUCGGAGCGCGAGGUGUCCACC---UCGGCCGAUGGCA-GUGCCAGGAA .((..(((((((((((.....)))))))))))..)).((((((((---(...))))))..((((.((((((....((((((....)))---))).)))))).))-)))))..... ( -56.40, z-score = -2.43, R) >droSec1.super_27 207791 108 + 799419 AACGGCACUGCUGUAGAGCUGCUGCAGCAGUGGAGUGGGUGUCCG---CGGUGCGGACAGGCUGGUGUCGGAGCGCGAGGUGUCCACC---UCGGCCGAUGGCA-GUGCCAGGAA .((..(((((((((((.....)))))))))))..)).((((((((---(...))))))..((((.((((((....((((((....)))---))).)))))).))-)))))..... ( -56.40, z-score = -2.51, R) >droYak2.chr3R 4924681 108 - 28832112 AACGGCACUGCUGUAGAGCUGCUGCAGCAGUGGAGUGGGUGUCCG---CGGGGCGGACAGGCUGGUGUCGGAUCGCGAAGUGUCCAGC---UCGGCCGAUGGCA-GUGCCAGGAA ...(((((((((((...(((((....)))))........((((((---(...)))))))((((((.((.(((.(((...)))))).))---)))))).))))))-)))))..... ( -52.30, z-score = -1.92, R) >droEre2.scaffold_4770 12217378 108 + 17746568 AACGGCACUGCUGUACAGCUGCUGCAGCAGUGGAGUGGGUGUCCG---CGGGGCGGACAGGCUGGUGUCGGAGCGCGAAGUGUCCACC---UCGGCCGAUGGCA-GUGCCAGGAA ...(((((((((((.(.(((((....)))))........((((((---(...)))))))((((((.((.(((.(((...)))))))).---)))))))))))))-)))))..... ( -52.00, z-score = -1.16, R) >droAna3.scaffold_12911 2457266 114 + 5364042 CACGGCGCUGCUGUAUAGCUGCUGAAGCAACGGAGUCGCAGCCCGACCAGACGCCGACAGACUGGUGUCGGAGCGCGAGGUGUCCACCGCAUCAGCUGGCGGCA-GAGCCAGGAA ...(((.(((((((......(((((.((.......((((.(((((((.....((((......))))))))).))))))(((....))))).)))))..))))))-).)))..... ( -50.20, z-score = -1.10, R) >dp4.chr2 15310465 102 - 30794189 GACGGCGCUGCUGUAGAGCUGCUGCAGCAGUGGAGUCUGGGCCCG---CGA---GGGCAGGCUGGUGUCGGAGCGCGAGGACUCCAUG---UCG---GGCGGCA-GCGCCAGGAA ...(((((((((((.(((((((....)))))((((((...((((.---...---))))...((.((((....)))).))))))))...---)).---.))))))-)))))..... ( -59.00, z-score = -3.29, R) >droPer1.super_0 6717227 102 + 11822988 GACGGCGCUGCUGUAGAGCUGCUGCAGCAGUGGAGUCUGGGCCCG---CGA---GGGCAGGCUGGUGUCGGAGCGCGAGGACUCCAUG---UCG---GGCGGCA-GCGCCAGGAA ...(((((((((((.(((((((....)))))((((((...((((.---...---))))...((.((((....)))).))))))))...---)).---.))))))-)))))..... ( -59.00, z-score = -3.29, R) >droWil1.scaffold_181108 1934960 90 + 4707319 CACCGCGCUGGUGUACAGCUGCUGCAGCAAU------------------------GGCAAAUUGCUAUCGGAACGCGAAUAUUCCAUGUCGUCUGCCGACAAGG-GCGCCAAAUA ..((((((((.....)))).))...((((((------------------------.....))))))...))...(((.....(((.(((((.....))))).))-))))...... ( -26.70, z-score = 0.44, R) >droMoj3.scaffold_6540 5329695 96 + 34148556 AACAGCGCUGCUGUACAGCUGCUGCAGUAGCUG---CGGUGUCUGAGCGCGCGACGAU------------GGGCGCGAGUUCUCCUGC---UCGGCCGGCGGCGCCAGCCAGUG- ....(((((((....(((((((....)))))))---(((...((((((((((......------------..))))((....))..))---)))))))))))))).........- ( -42.70, z-score = 1.12, R) >droGri2.scaffold_15074 4067280 99 - 7742996 CAACGCGCUGCUGUAGAUCUGUUGCAGCAGCAA---CGGCGUUG----UGGCACGCGAAGGCUGCU-----GGCUUGAGCAUUCAGUU---GGUGUCGGCGGCGCUAGCCACUG- .(((((((((((((((.....))))))))))..---..)))))(----((((..((....((((((-----(((.(.(((.....)))---.).)))))))))))..)))))..- ( -46.00, z-score = -1.23, R) >droVir3.scaffold_13047 13035029 105 - 19223366 CACGGCGCUGCUAUAUAGCUGCUGCAGUAG------CGGCGUCUGGGCGCGCGACGACUGGCUGUUGUCGGAGCGGGAGUUCUCCUGC---UCGGCCGGCGGUGCCAGCCAUUG- ...(((((((((...(((.((((((....)------))))).)))(((...(((((((.....)))))))((((((((....))))))---)).))))))))))))........- ( -57.40, z-score = -2.67, R) >consensus AACGGCGCUGCUGUAGAGCUGCUGCAGCAGUGGAGUCGGUGUCCG___CGGCGCGGACAGGCUGGUGUCGGAGCGCGAGGUGUCCACC___UCGGCCGACGGCA_GUGCCAGGAA ....(((((((((((.......))))))))).........(((............)))................))........................(((....)))..... (-15.17 = -15.70 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:33 2011