
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 16,109,899 – 16,110,000 |
| Length | 101 |
| Max. P | 0.776411 |

| Location | 16,109,899 – 16,110,000 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 63.48 |
| Shannon entropy | 0.72293 |
| G+C content | 0.36918 |
| Mean single sequence MFE | -13.47 |
| Consensus MFE | -6.56 |
| Energy contribution | -6.52 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776411 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 16109899 101 + 27905053 AGUUGGCAACAGCAUAAAAUACAACGUAAAAAU-GUCAGAAAAAAUAUGCACAGA-------AAAAUCACUGCAAACAAAUUU-ACAACGCUAACUGCACUCCCCCGCUA---- (((((((....(((((.......((((....))-)).........))))).(((.-------.......)))...........-.....)))))))((........))..---- ( -14.69, z-score = -0.97, R) >droSim1.chr3R 22144193 103 - 27517382 AGUUGGCAACAGCAUAAAAUACAACGUAAAAAU-GUCAGAAAAAAUAUGCACAGA-------AAAAUCACAGCAAACAAAUUC-ACAACGCUAACUGCACUCCCCCACUAUC-- (((((((....(((((.......((((....))-)).........)))))...((-------....))...............-.....)))))))................-- ( -12.59, z-score = -1.02, R) >droSec1.super_27 170229 103 - 799419 AGUUGGCAACAGCAUAAAAUACAACGUAAAAAU-GUCAGAAAAAAUAUGCACAGA-------AAAAUCACAGCAAACAAAUUC-ACAACGCUAACUGCACUCCCCCACUAUC-- (((((((....(((((.......((((....))-)).........)))))...((-------....))...............-.....)))))))................-- ( -12.59, z-score = -1.02, R) >droYak2.chr3R 4885288 100 + 28832112 AGUUGGCAACAGCAUAAAAUACAACGUAAAAAU-GGAUGAAAAAAUGUGCACAGA-------AAAAUCACAGCAAACAAAUUC-ACAACGCUAACUGCACUCCCCCGCU----- .((((....))))....................-(((((((......(((...((-------....))...)))......)))-)....((.....))..)))......----- ( -13.60, z-score = 0.18, R) >droAna3.scaffold_13340 2646078 105 + 23697760 AGUUGGCAACAGCAUAAAAUUCAACGUAAAAAUAGAGAGAAAAAAUUCAAAGAAAGGCACAUAAAAUCACAGCAAACAAAUUC-ACUACGCUAACUGCACUCCUAC-------- .((((....))))...................(((.(((.................((.............))..........-.....((.....)).)))))).-------- ( -12.72, z-score = -0.96, R) >dp4.chr2 3216297 91 - 30794189 AGUUGGCAACAACAUAAAGUACUUAACAAAAA-----AAAAAAAAAAU------------------UCAUUGCAAACAAAUUCUGAAAUGCUAACUGCACUCGCUUGCCUGCUC .((((....))))...................-----...........------------------.....(((..(((....(((..(((.....))).))).)))..))).. ( -11.60, z-score = -0.18, R) >droPer1.super_6 3241697 88 - 6141320 AGUUGGCAACAACAUAAAGUACUUAACAAAAA-----AAAAAAA---U------------------UCAUUGCAAACAAAUUCUGAAAUGCUAACUGCACUCGCUUGCCUGCUC .((((....))))...................-----.......---.------------------.....(((..(((....(((..(((.....))).))).)))..))).. ( -11.60, z-score = -0.17, R) >droWil1.scaffold_181089 1636957 83 - 12369635 -----------------------UUGUGACGAGAGAGAGAAGGACGCCAUGCGUUG------GGAACAACAAAAAAAUCACAGCAUUAUGCUAACUGCAAUUGCAUGAGCUG-- -----------------------......(....)..........(((((((((((------....))))..................(((.....)))...))))).))..-- ( -18.40, z-score = -0.92, R) >consensus AGUUGGCAACAGCAUAAAAUACAACGUAAAAAU_GUCAGAAAAAAUAUGCACAGA_______AAAAUCACAGCAAACAAAUUC_ACAACGCUAACUGCACUCCCCCGCUA_C__ .((((....))))............................................................................((.....))................ ( -6.56 = -6.52 + -0.03)

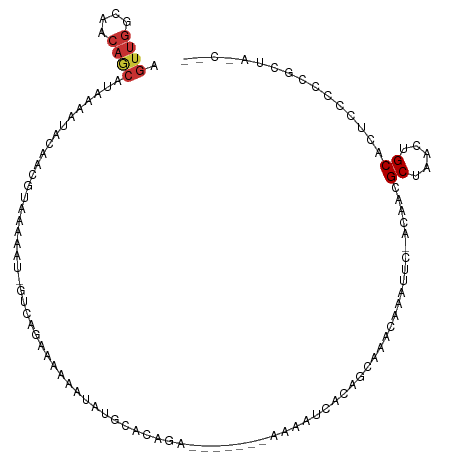

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:29:29 2011